Mapping of a myosin-binding domain and a regulatory phosphorylation site in M-protein, a structural protein of the sarcomeric M band - PubMed (original) (raw)
Mapping of a myosin-binding domain and a regulatory phosphorylation site in M-protein, a structural protein of the sarcomeric M band
W M Obermann et al. Mol Biol Cell. 1998 Apr.
Free PMC article
Abstract
The myofibrils of cross-striated muscle fibers contain in their M bands cytoskeletal proteins whose main function seems to be the stabilization of the three-dimensional arrangement of thick filaments. We identified two immunoglobin domains (Mp2-Mp3) of M-protein as a site binding to the central region of light meromyosin. This binding is regulated in vitro by phosphorylation of a single serine residue (Ser76) in the immediately adjacent amino-terminal domain Mp1. M-protein phosphorylation by cAMP-dependent kinase A inhibits binding to myosin LMM. Transient transfection studies of cultured cells revealed that the myosin-binding site seems involved in the targeting of M-protein to its location in the myofibril. Using the same method, a second myofibril-binding site was uncovered in domains Mp9-Mp13. These results support the view that specific phosphorylation events could be also important for the control of sarcomeric M band formation and remodeling.
Figures
Figure 1
Summary diagram giving a schematic representation of the domain organizations for M-protein and for myosin. The presentation emphasizes the modular construction of M-protein from repetitive Ig cII (cross-hatched rectangles) and fibronectin type III repeats (shaded rectangles) interspersed by unique sequence stretches of varying length. Recombinant constructs used for mapping of binding sites, transfection of cultured myoblasts, and phosphorylation assays are marked by brackets. The location of the most prominent proteolytic fragments of myosin is given in the lower panel above the myosin sketch. Brackets indicate the borders of the recombinant LMM constructs used in this study.
Figure 2
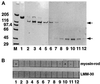
Binding of M-protein and its proteolytic 45-kDa fragment to myosin rod. (A) SDS-PAGE analysis (4–14%) of M-protein purified from bovine skeletal muscle (lane 1) and gel filtration fractions of an endoproteinase Asp-N digest of M-protein (see MATERIALS AND METHODS) (lanes 2–12). M = molecular mass standards in kilodaltons. (B) M-protein (1) and corresponding fractions of the M-protein digest (2–12) spotted on nitrocellulose filters after incubation with biotinylated myosin-rod or LMM 30. Note that only native M-protein (1) and its 45-kDa fragment (comprising domains Mp2–Mp4; fractions 9–11) show binding to myosin rod, while the proteolytic 110-kDa fragment of M-protein (comprising domains Mp5–Mp13; lanes 2–6) does not bind to myosin rod (the positions of these proteolytic fragments are indicated by arrows). Note also that both M-protein fragments do not bind to LMM30. Thus myosin rod binding of M-protein requires domains Mp2–Mp4.
Figure 3
Binding of recombinant M-protein fragments to proteolytic and recombinant derivatives of myosin. (A) SDS-PAGE analysis (6–20% gradient gels) of purified recombinant M-protein fragments: Mp2 (lane 1), Mp2–Mp3 (lane 2), Mp3 (lane 3), and Mp4 (lane 4). M = molecular mass standards in kilodaltons. For domain structure of M-protein see Figure 1. (B) Results of binding assays. The same amounts (ca. 1 μg) of myosin rod, proteolytic LMM, LMM 75, LMM 59, LMM 50, LMM 30, LMM 50–75, and ovalbumin, serving as a control, were spotted on nitrocellulose filters and overlaid with increasing concentrations (1 = 0.5 μM, 2 = 1.5 μM, 3 = 4.5 μM) of M-protein fragments Mp2 (a), Mp2 to Mp3 (b), Mp3 (c), and Mp4 (d). (e) Control without protein in the overlay buffer. Binding of M-protein fragments carrying the carboxy-terminal EEF-tag was detected with monoclonal antibody YL1/2, which specifically recognizes this tag. Note the specific binding of M-protein fragment Mp2–Mp3 to all myosin fragments that contain the central portion of LMM (myosin heavy chain residues 1506–1674) but not to LMM30 and LMM50–75.
Figure 4
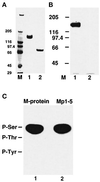
Phosphorylation of native M-protein and phosphoamino acid analysis of M-protein and a recombinant Mp1–5 fragment. (A) M-protein from bovine skeletal muscle (lane 1) and a limited digest with trypsin (lane 2), analyzed on 4–12% SDS-PAGE (see MATERIALS AND METHODS). (B) The corresponding autoradiograph of the samples shown in panel A after incubation with PKA in the presence of [γ-32P] ATP shows that M-protein (lane 1) is readily phosphorylated while its tryptic fragment (Mp6– Mp13) is not (lane 2). M = molecular mass standards in kilodaltons. (C) Phosphoamino acid analysis of native M-protein from bovine skeletal muscle (lane 1) and the recombinant M-protein fragment Mp1–Mp5 (lane 2) after phosphorylation with PKA. Positions of marker amino acids are indicated. Clearly, phosphorylation occurs in both samples exclusively on serine residues.
Figure 5
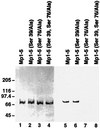
In vitro phosphorylation of mutant recombinant M-protein fragments. Purified EEF-tagged Mp1–Mp5 fragments were phosphorylated in vitro by PKA and 32P-labeled ATP and run on two identical 4–10% gradient polyacrylamide gels. One gel was blotted to nitrocellulose and stained with the antibody recognizing the EEF-tag and a peroxidase-coupled secondary antibody (lanes 1–4), while the second gel was dried and autoradiographed (lanes 5–8). Lanes 1–4 contain approximately identical amounts of Mp1–Mp5 (lane 1), Mp1–Mp5 (Ser39/Ala) (lane 2), Mp1–Mp5 (Ser76/Ala) (lane 3), and Mp1–Mp5 (Ser39, 76/Ala) (lane 4). Lanes 5–8 show that Mp1–Mp5 and its (Ser39/Ala) mutant are phosphorylated, while phosphorylation of both mutants containing the Ser76/Ala mutation (lanes 7 and 8) is almost completely abolished. Thus Ser76 is the PKA phosphorylation site of M-protein.
Figure 6
Interaction of unphosphorylated and PKA phosphorylated M-protein with proteolytic and recombinant derivatives of myosin. The same amounts (1 μg) of myosin-rod, proteolytic LMM, LMM75, LMM59, LMM50, LMM30, LMM50–75, and ovalbumin, serving as a control, were spotted on nitrocellulose filters and overlayed with increasing concentrations (1 = 0.1 μM, 2 = 0.3 μM, 3 = 1.0 μM) of unphosphorylated (a) and phosphorylated (b) M-protein. (c) is a control without protein. Binding of M-protein was detected using the monoclonal M-protein antibody MpAA280 (see Obermann, et al., 1996). Note that phosphorylation of M-protein almost completely abolished binding to myosin derivatives.
Figure 7
Expression of recombinant M-protein fragments in transiently transfected BHK-21/C13 cells. BHK-21/C13 cells transfected with constructs encoding Mp2 (A and B), Mp3 (C and D), or Mp2–Mp3 (E–H) were double stained with T7-tag antibody to localize the recombinant M-protein fragment (A, C, E, and G) and a titin antibody (B, D, F, and H). (A– D) Examples of transfected cells that contain large amounts of diffusely distributed recombinant protein (A and C) and titin, which appears in a normal staining pattern (arrowheads in B and D). The expressed Mp2–Mp3 construct either disrupts MLS, which then results in the appearance of numerous cytoplasmic aggregates containing both the recombinant protein and titin (arrowheads in E and F), or colocalizes with MLS (arrowheads in G and H). For details see RESULTS. Magnification, 1050×.
Figure 8
Expression of recombinant M-protein fragments in differentiating C2C12 cells. C2C12 cells transfected with constructs encoding T7-tagged Mp2–Mp3 (A–D) or Mp9–Mp13 (E) were allowed to differentiate for 2 (E) or 6 (A to D) d, respectively. Subsequently they were stained with T7-tag antibody (A, C, and E) and tetramethylrhodamine-5-isothiocyanate-labeled phalloidin (B) or MpAA259 (D) followed by secondary antibody. Note that most of the expressed Mp2–Mp3 polypeptide associates with myofibrils (arrowheads in A–D). Transfection with Mp9–Mp13 leads to expression of a polypeptide that binds to myofibrils in a periodic manner (arrowheads in E). For details see RESULTS. Magnification, 1050×.
Similar articles
- The structure of the sarcomeric M band: localization of defined domains of myomesin, M-protein, and the 250-kD carboxy-terminal region of titin by immunoelectron microscopy.
Obermann WM, Gautel M, Steiner F, van der Ven PF, Weber K, Fürst DO. Obermann WM, et al. J Cell Biol. 1996 Sep;134(6):1441-53. doi: 10.1083/jcb.134.6.1441. J Cell Biol. 1996. PMID: 8830773 Free PMC article. - Transient association of titin and myosin with microtubules in nascent myofibrils directed by the MURF2 RING-finger protein.
Pizon V, Iakovenko A, Van Der Ven PF, Kelly R, Fatu C, Fürst DO, Karsenti E, Gautel M. Pizon V, et al. J Cell Sci. 2002 Dec 1;115(Pt 23):4469-82. doi: 10.1242/jcs.00131. J Cell Sci. 2002. PMID: 12414993 - Dynamic regulation of sarcomeric actin filaments in striated muscle.
Ono S. Ono S. Cytoskeleton (Hoboken). 2010 Nov;67(11):677-92. doi: 10.1002/cm.20476. Cytoskeleton (Hoboken). 2010. PMID: 20737540 Free PMC article. Review. - Connectin/titin, giant elastic protein of muscle.
Maruyama K. Maruyama K. FASEB J. 1997 Apr;11(5):341-5. doi: 10.1096/fasebj.11.5.9141500. FASEB J. 1997. PMID: 9141500 Review.
Cited by
- Myofibrillogenesis regulator 1 induces hypertrophy by promoting sarcomere organization in neonatal rat cardiomyocytes.
Wang X, Liu X, Wang S, Luan K. Wang X, et al. Hypertens Res. 2012 Jun;35(6):597-603. doi: 10.1038/hr.2011.228. Epub 2012 Mar 15. Hypertens Res. 2012. PMID: 22418241 Free PMC article. - Thick filament assembly occurs after the formation of a cytoskeletal scaffold.
Van der Ven PF, Ehler E, Perriard JC, Fürst DO. Van der Ven PF, et al. J Muscle Res Cell Motil. 1999 Aug;20(5-6):569-79. doi: 10.1023/a:1005569225773. J Muscle Res Cell Motil. 1999. PMID: 10555075 - Synaptopodin-2 Isoforms Have Specific Binding Partners and Display Distinct, Muscle Cell Type-Specific Expression Patterns.
Lohanadan K, Assent M, Linnemann A, Schuld J, Heukamp LC, Krause K, Vorgerd M, Reimann J, Schänzer A, Kirfel G, Fürst DO, Van der Ven PFM. Lohanadan K, et al. Cells. 2023 Dec 30;13(1):85. doi: 10.3390/cells13010085. Cells. 2023. PMID: 38201288 Free PMC article. - The role of the M-band myomesin proteins in muscle integrity and cardiac disease.
Lamber EP, Guicheney P, Pinotsis N. Lamber EP, et al. J Biomed Sci. 2022 Mar 7;29(1):18. doi: 10.1186/s12929-022-00801-6. J Biomed Sci. 2022. PMID: 35255917 Free PMC article. Review. - Novel mutations widen the phenotypic spectrum of slow skeletal/β-cardiac myosin (MYH7) distal myopathy.
Lamont PJ, Wallefeld W, Hilton-Jones D, Udd B, Argov Z, Barboi AC, Bonneman C, Boycott KM, Bushby K, Connolly AM, Davies N, Beggs AH, Cox GF, Dastgir J, DeChene ET, Gooding R, Jungbluth H, Muelas N, Palmio J, Penttilä S, Schmedding E, Suominen T, Straub V, Staples C, Van den Bergh PY, Vilchez JJ, Wagner KR, Wheeler PG, Wraige E, Laing NG. Lamont PJ, et al. Hum Mutat. 2014 Jul;35(7):868-79. doi: 10.1002/humu.22553. Epub 2014 May 21. Hum Mutat. 2014. PMID: 24664454 Free PMC article.
References
- Andersson S, Davis DN, Dahlbäck H, Jörnval H, Russel DW. Cloning, structure and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264:8222–8229. - PubMed
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short Protocols in Molecular Biology. 3rd ed. New York: Wiley and Sons, Inc.; 1995.
- Bähler M, Wallimann T, Eppenberger HM. Myofibrillar M-band proteins represent constituents of native thick filaments, frayed filaments and bare zone assemblages. J Muscle Res Cell Motil. 1985;6:783–800. - PubMed
- Carlsson E, Grove BK, Wallimann T, Eppenberger HM, Thornell L-E. Myofibrillar M-band proteins in rat skeletal muscles during development. Histochemistry. 1990;95:27–35. - PubMed
- Casnellie JE. Assay of protein kinases using peptides with basic residues for phosphocellulose binding. Methods Enzymol. 1991;200:115–120. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases