Arp2/3 complex from Acanthamoeba binds profilin and cross-links actin filaments - PubMed (original) (raw)
Arp2/3 complex from Acanthamoeba binds profilin and cross-links actin filaments
R D Mullins et al. Mol Biol Cell. 1998 Apr.
Free PMC article
Abstract
The Arp2/3 complex was first purified from Acanthamoeba castellanii by profilin affinity chromatography. The mechanism of interaction with profilin was unknown but was hypothesized to be mediated by either Arp2 or Arp3. Here we show that the Arp2 subunit of the complex can be chemically cross-linked to the actin-binding site of profilin. By analytical ultracentrifugation, rhodamine-labeled profilin binds Arp2/3 complex with a Kd of 7 microM, an affinity intermediate between the low affinity of profilin for barbed ends of actin filaments and its high affinity for actin monomers. These data suggest the barbed end of Arp2 is exposed, but Arp2 and Arp3 are not packed together in the complex exactly like two actin monomers in a filament. Arp2/3 complex also cross-links actin filaments into small bundles and isotropic networks, which are mechanically stiffer than solutions of actin filaments alone. Arp2/3 complex is concentrated at the leading edge of motile Acanthamoeba, and its localization is distinct from that of alpha-actinin, another filament cross-linking protein. Based on localization and actin filament nucleation and cross-linking activities, we propose a role for Arp2/3 in determining the structure of the actin filament network at the leading edge of motile cells.
Figures
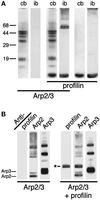
Figure 1
EDC with NHS cross-links Acanthamoeba profilin to the Arp2 subunit of the Arp2/3 complex. (A) SDS-PAGE of proteins cross-linked with EDC/NHS and stained with Coomassie blue (cb) or immunoblotted (ib) with monoclonal antiprofilin antibodies. Samples, as indicated by the horizontal bars: 3 μM Arp2/3 complex, Arp2/3 plus 20 μM profilin-II, profilin-II alone. (B) Immunoblot of Arp2/3 complex and profilin-II cross-linked with EDC/NHS and probed with antibodies against profilin (lanes 1 and 4), Arp2 (lanes 2 and 5), and Arp3 (lanes 3 and 6). Lanes 1–3, 3 μM Arp2/3 complex; lanes 4–6, 3 μM Arp2/3 and 20 μM profilin-II. Asterisk marks the two profilin/Arp2 cross-linked products. Identical results were obtained with Acanthamoeba profilin-I. Conditions: Cross-linking was carried out for 1 h at 24°C with 5 mM EDC and 5 mM NHS. Buffer: 150 mM NaCl, 0.2 mM MgCl2, 0.1 mM EGTA, 0.2 mM ATP, 1 mM DTT, 10 mM imidazole, pH 7.5. Arp2/3 complex was purified by profilin:poly-L-proline affinity chromatography.
Figure 2
Arp2 binds at or near the actin-binding site on profilin. Immunoblot of SDS-PAGE of profilin-II cross-linked to Arp2/3 complex with EDC/NHS and probed with four epitope-mapped, monoclonal anti-profilin antibodies, P4, P5, P6, and P7. Antibodies P4 and P6 bind free profilin and profilin cross-linked to actin by EDC/NHS. Both also react with the two Arp2-profilin cross-linked products. P5 and P7 bind free profilin but not profilin cross-linked to actin and do not bind the smaller of the two Arp2-profilin cross-linked products. Conditions as in Figure 1.
Figure 3
Analytical ultracentrifugation analysis of profilin-II binding to (A) actin and (B) Arp2/3 complex. Samples were centrifuged to equilibrium at 20,000 rpm at 24°C in an An60ti rotor in a Beckman model XL-A analytical ultracentrifuge. (A) Sedimentation equilibrium distributions of 5.3 μM rhodamine-labeled N58C Acanthamoeba profilin-II in the absence (triangles) and presence (circles) of Acanthamoeba actin. Left panel, 5 μM monomeric actin. Right panel, 2.5 μM monomeric actin. Solid lines are fits to the data assuming a Kd of 0.9 μM. Residuals of the fit are shown in the lower panels. (B) Sedimentation equilibrium distributions of 5.3 μM rhodamine-labeled profilin with (diamonds) and without (circles) Arp2/3 complex. Left panel, 8 μM Arp2/3 complex; right panel, 4 μM Arp2/3 complex. Solid lines are least squares fits of a two-component model with a Kd of 7.4 μM. Residuals are shown in the lower panels. Conditions were as follows. Buffer: 150 mM NaCl, 0.2 mM MgCl2, 0.1 mM ATP, 10 mM imidazole, pH 7.5. Temperature: 25°C. Arp2/3 complex was purified by profilin:poly-L-proline affinity chromatography.
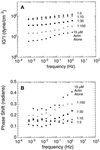
Figure 4
Rheological properties of actin filaments in the presence of Arp2/3 complex. (A) Frequency dependence of complex modulus, ‖G*‖. (B) Frequency dependence of the phase shift. Samples contained 15 μM rabbit skeletal muscle actin in the absence (•) or presence of Arp2/3 complex at 0.1 μM (○), 0.5 μM (▪), 1.5 μM (□), or 3 μM (▴). Conditions were as follows. Cone and plate rheometer in the small amplitude (strain ≤ 2%), forced oscillation mode. Buffer: 50 mM KCl, 10 mM imidazole, pH 7.0, 1 mM EGTA, and 1 mM MgCl2_._ Arp2/3 complex was purified by conventional ion exchange chromatography.
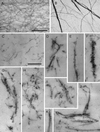
Figure 5
Electron micrographs of negatively stained or thin-sectioned actin filaments and Arp2/3-induced filament bundles. (A and B) Negative stained samples. (C–K) Thin sectioned samples. Actin alone (A, C). Actin with Arp2/3 complex (B, D–K). Bar in panels A and B is 395 nm. Bar in panels C–K is 395 nm. Conditions were as follows. Negative stained samples: samples of either 3 μM Acanthamoeba actin alone or 3 μM actin and 0.5 μM Arp2/3 complex were adsorbed to carbon-coated electron microscope grids and negatively stained with 1% uranyl formate (see MATERIALS AND METHODS). Thin sectioned samples: 10 μM Acanthamoeba actin was polymerized in the absence or presence of 1 μM Arp2/3 complex. Samples were fixed, stained, embedded in Epon, and thin-sectioned (see MATERIALS AND METHODS). Arp2/3 complex in the negatively stained samples was purified by profilin:poly-L-proline affinity chromatography and in the thin-sectioned samples by conventional ion exchange chromatography.
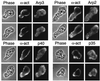
Figure 6
Localization of Arp2/3 complex and α-actinin in Acanthamoeba castellanii by indirect immunofluorescence. Phase: phase contrast micrographs of individual adherent amoebas oriented with their leading edges pointing toward the upper right. α-act: distribution of α-actinin. Arp3/Arp2/p40/p35: individual distributions of four different subunits of the Arp2/3 complex.
Similar articles
- Structure, subunit topology, and actin-binding activity of the Arp2/3 complex from Acanthamoeba.
Mullins RD, Stafford WF, Pollard TD. Mullins RD, et al. J Cell Biol. 1997 Jan 27;136(2):331-43. doi: 10.1083/jcb.136.2.331. J Cell Biol. 1997. PMID: 9015304 Free PMC article. - Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins.
Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Blanchoin L, et al. Nature. 2000 Apr 27;404(6781):1007-11. doi: 10.1038/35010008. Nature. 2000. PMID: 10801131 - Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks.
Blanchoin L, Pollard TD, Mullins RD. Blanchoin L, et al. Curr Biol. 2000 Oct 19;10(20):1273-82. doi: 10.1016/s0960-9822(00)00749-1. Curr Biol. 2000. PMID: 11069108 - Actin-based motility as a self-organized system: mechanism and reconstitution in vitro.
Carlier MF, Wiesner S, Le Clainche C, Pantaloni D. Carlier MF, et al. C R Biol. 2003 Feb;326(2):161-70. doi: 10.1016/s1631-0691(03)00067-2. C R Biol. 2003. PMID: 12754935 Review. - Control of actin assembly and disassembly at filament ends.
Cooper JA, Schafer DA. Cooper JA, et al. Curr Opin Cell Biol. 2000 Feb;12(1):97-103. doi: 10.1016/s0955-0674(99)00062-9. Curr Opin Cell Biol. 2000. PMID: 10679358 Review.
Cited by
- Profilin enhances Cdc42-induced nucleation of actin polymerization.
Yang C, Huang M, DeBiasio J, Pring M, Joyce M, Miki H, Takenawa T, Zigmond SH. Yang C, et al. J Cell Biol. 2000 Sep 4;150(5):1001-12. doi: 10.1083/jcb.150.5.1001. J Cell Biol. 2000. PMID: 10973991 Free PMC article. - A role for myosin-I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex.
Evangelista M, Klebl BM, Tong AH, Webb BA, Leeuw T, Leberer E, Whiteway M, Thomas DY, Boone C. Evangelista M, et al. J Cell Biol. 2000 Jan 24;148(2):353-62. doi: 10.1083/jcb.148.2.353. J Cell Biol. 2000. PMID: 10648568 Free PMC article. - Branched actin cortices reconstituted in vesicles sense membrane curvature.
Baldauf L, Frey F, Arribas Perez M, Idema T, Koenderink GH. Baldauf L, et al. Biophys J. 2023 Jun 6;122(11):2311-2324. doi: 10.1016/j.bpj.2023.02.018. Epub 2023 Feb 17. Biophys J. 2023. PMID: 36806830 Free PMC article. - A mutant of Arp2p causes partial disassembly of the Arp2/3 complex and loss of cortical actin function in fission yeast.
Morrell JL, Morphew M, Gould KL. Morrell JL, et al. Mol Biol Cell. 1999 Dec;10(12):4201-15. doi: 10.1091/mbc.10.12.4201. Mol Biol Cell. 1999. PMID: 10588653 Free PMC article. - Relating interactions between neurofilaments to the structure of axonal neurofilament distributions through polymer brush models.
Kumar S, Yin X, Trapp BD, Hoh JH, Paulaitis ME. Kumar S, et al. Biophys J. 2002 May;82(5):2360-72. doi: 10.1016/S0006-3495(02)75581-1. Biophys J. 2002. PMID: 11964226 Free PMC article.
References
- Bubb MR, Lewis MS, Korn ED. Actobindin binds with high affinity to a covalently cross-linked actin dimer. J Biol Chem. 1994;269:25587–25591. - PubMed
- Cooper JA, Pollard TD. Effects of capping protein on the kinetics of actin polymerization. Biochemistry. 1985;24:793–799. - PubMed
- Ferry JD. Viscoelestic Properties of Polymers. 3rd ed. New York: John Wiley and Sons; 1980.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous