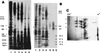Cell wall and secreted proteins of Candida albicans: identification, function, and expression - PubMed (original) (raw)
Review
Cell wall and secreted proteins of Candida albicans: identification, function, and expression
W L Chaffin et al. Microbiol Mol Biol Rev. 1998 Mar.
Abstract
The cell wall is essential to nearly every aspect of the biology and pathogenicity of Candida albicans. Although it was initially considered an almost inert cellular structure that protected the protoplast against osmotic offense, more recent studies have demonstrated that it is a dynamic organelle. The major components of the cell wall are glucan and chitin, which are associated with structural rigidity, and mannoproteins. The protein component, including both mannoprotein and nonmannoproteins, comprises some 40 or more moieties. Wall proteins may differ in their expression, secretion, or topological location within the wall structure. Proteins may be modified by glycosylation (primarily addition of mannose residues), phosphorylation, and ubiquitination. Among the secreted enzymes are those that are postulated to have substrates within the cell wall and those that find substrates in the extracellular environment. Cell wall proteins have been implicated in adhesion to host tissues and ligands. Fibrinogen, complement fragments, and several extracellular matrix components are among the host proteins bound by cell wall proteins. Proteins related to the hsp70 and hsp90 families of conserved stress proteins and some glycolytic enzyme proteins are also found in the cell wall, apparently as bona fide components. In addition, the expression of some proteins is associated with the morphological growth form of the fungus and may play a role in morphogenesis. Finally, surface mannoproteins are strong immunogens that trigger and modulate the host immune response during candidiasis.
Figures
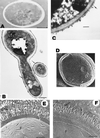
FIG. 1
Cell wall structure. (A) Transmission electron micrograph of a section of a C. albicans cell prepared by freeze-substitution, showing the cell wall as a thick, electron-dense, homogeneous structure. The presence of distinct layers was not evident in this preparation. Bar, 1 μm. Reprinted from reference with permission of the publisher. (B) Thin sections of cells treated with gold-conjugated concanavalin A, showing an intense labeling with gold particles of the external wall surface. The surface exhibits a fibrillar appearance (arrows), suggesting that concanavalin A-reactive cell wall components, i.e., mannoproteins, are particularly abundant at the most external wall layers. The remaining wall structure also appeared as a homogenous structure in this transmission electron micrograph. Bar, 0.5 μm. Reprinted from reference with permission of the American Society for Microbiology. (C) Other procedures for transmission electron microscopy examination of thin sections of C. albicans cells revealed more clearly the presence of an outer floccular layer (arrow) and showed that the remaining cell wall structure is not homogeneous and that some layering exists. Bar, 200 nm. Reprinted from reference with permission of the publisher. (D to F) Complexity of the wall ultrastructure and presence of distinct layers in the cell wall of C. albicans as revealed by different scanning electron microscopy-based procedures such as cryo-scanning electron microscopy (D) and freeze-fracture, freeze-etch analysis (E and F). The presence of well-ordered, regularly arranged, radiating fibrils in the outer layer is particularly evident in the micrographs shown in panels E (hydrophilic cells) and F (hydrophobic cells). Bar, 0.3 μm in both panels. Panel D reprinted from reference with permission of the publisher. Panels E and F reprinted from reference with permission of the publisher.
FIG. 2
Polypeptide composition of cell wall extracts and whole protoplast homogenates from C. albicans as revealed by different electrophoretic techniques (A and B). C. albicans cells from which samples were obtained were incubated in the presence of 14C-labelled protein hydrolysate and subsequently tagged with biotin. Double-labelled cell wall proteins and glycoproteins were extracted from intact blastoconidia (lanes 1 and 3) and germinated blastoconidia (lanes 2 and 4) by sequential treatment with βME (lanes 1 and 2) and digestion with Zymolyase 20T (lanes 3 and 4). Samples of protoplast homogenates from blastoconidia and germinated blastoconidia were run in lanes 5 and 6, respectively. Polypeptides were separated by SDS-PAGE and detected by fluorography (A) or transferred to nitrocellulose and detected with an avidin-peroxidase conjugate (B; lane S in this panel shows a mixture of prestained molecular weight standards run in parallel). Numbers and letters are used to identify and compare bands detected in the cell wall extracts by the different experimental procedures used. Although qualitative differences were observed (i.e., some polypeptides exhibited a strong radioactive label but were weakly biotinylated [band 6; star]), surface labelling of cells with biotin appeared to be a suitable technique to detect proteins in the wall of C. albicans. Thus, from the complex polypeptide pattern found in the protoplast homogenate samples as revealed by fluorography (A, lanes 5 and 6), only few species were labelled with biotin, indicating that most proteins released by βME and Zymolyase from intact cells are bona fide cell wall components. Brackets indicate a cluster of bands within a molecular weight range where many candidal moieties that represent receptors for host ligands have been identified (see the text). Reprinted from reference with permission of the American Society for Microbiology. (C) The complexity of the polypeptide pattern of the cell wall extracts was clearly evidenced when analysis was performed by two-dimensional PAGE and silver staining (the polypeptide pattern shown corresponds to the βME extract from blastoconidia). Reprinted from reference with permission of the American Society for Microbiology.
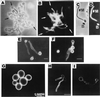
FIG. 3
Surface expression of cell wall proteins. Phase-contrast (A and C) and immunofluorescence (B, D, and E to I) of C. albicans blastoconidia (B) and mycelial filaments (M) reacted with different polyclonal and monoclonal antibody preparations raised to protein and glycoprotein cell wall constituents. Some antibodies recognized antigens that appeared to be specific for or preferentially expressed in germ tubes (A and D) or blastoconidia (E and F). Arrows in panels A to D point to the location of mother blastoconidia (A and C) that exhibited no fluorescence (B and D). Some antigens appeared to be homogenously distributed on the surface of mycelial filaments (B and D) or blastoconidia (G). However, patches of greater fluorescence intensity were observed with other antisera preparations (H and I), suggesting that antigens recognized by such antisera were heterogenously and randomly distributed within the cell wall structure. The pictures in panels B and D to G are from standard immunofluorescence microscopy observations. Panels H and I show single-focal-plane sections of different cells obtained by confocal fluorescence microscopy and associated software. Bar, 10 μm (except for panel G, which is 1.25 μm). Panels A to D reprinted from reference with permission of the American Society for Microbiology. Panels E and F reprinted from reference with permission of the American Society for Microbiology. Panel G reprinted from reference with permission of the American Society for Microbiology. Panels H and I reprinted from reference with permission of the American Society for Microbiology.
FIG. 4
Schematic diagram of the cell wall (CW) structure of C. albicans, showing the presence of different layers enriched in particular components. The microfibrillar polymers of β-glucans (β-g, ) and chitin (c,  ) appear to be more heavily concentrated in the inner cell wall domains; β-glucan–chitin complexes that appear to be formed by glycosidic linkages between both polymers will be located adjacent to the plasma membrane (PM) and the periplasmic space (ps). Proteins and glyco(manno)proteins (gp) appear to be dominant in the outermost cell wall layer, although they are also distributed through the entire wall structure. Once secreted through the plasma membrane, some protein and glycoproteins species will remain at the periplasmic space, possibly playing enzymatic roles (▵); some others will establish functional (i.e., β-glucanases [▾]) or structural covalent associations with β-glucans and possibly also with chitin (•—•) adjacent to the plasma membrane; and, finally, other moieties will constitute the most external layer, where the different molecular entities may be homogeneously (□) or heterogeneously (fimbriae, cluster of receptor-like molecules, etc. [○]) distributed or specifically released (i.e., extracellular enzymes) to the extracellular medium (EM) (•, ▪). Proteins and glycoprotein species in the outermost wall layer (□, ○) may establish different types of covalent (disulfide linkages) and noncovalent (hydrophobic and hydrogen ionic bonds) interactions. During their passage through the wall from the plasma membrane and periplasmic space to the outermost cell wall layers (□, ○) and possibly the extracellular environment (•, ▪), proteins and glycoproteins are most likely to be in equilibrium with other proteinaceous constituents, thus contributing, at least from a functional point of view, to the cell wall layering. In any case, protein and glycoprotein species other than those specifically secreted to the exocellular medium may also be released to such locations by dying (lysed) cells or as a consequence of unbalanced processes of synthesis and degradation of the cell wall structure, required for wall expansion during cell growth. To simplify the scheme, some aspects such as possible interactions of cell wall components with the plasma membrane and proteins retained in the cell wall, apparently by either covalent or noncovalent linkages, are not depicted.
) appear to be more heavily concentrated in the inner cell wall domains; β-glucan–chitin complexes that appear to be formed by glycosidic linkages between both polymers will be located adjacent to the plasma membrane (PM) and the periplasmic space (ps). Proteins and glyco(manno)proteins (gp) appear to be dominant in the outermost cell wall layer, although they are also distributed through the entire wall structure. Once secreted through the plasma membrane, some protein and glycoproteins species will remain at the periplasmic space, possibly playing enzymatic roles (▵); some others will establish functional (i.e., β-glucanases [▾]) or structural covalent associations with β-glucans and possibly also with chitin (•—•) adjacent to the plasma membrane; and, finally, other moieties will constitute the most external layer, where the different molecular entities may be homogeneously (□) or heterogeneously (fimbriae, cluster of receptor-like molecules, etc. [○]) distributed or specifically released (i.e., extracellular enzymes) to the extracellular medium (EM) (•, ▪). Proteins and glycoprotein species in the outermost wall layer (□, ○) may establish different types of covalent (disulfide linkages) and noncovalent (hydrophobic and hydrogen ionic bonds) interactions. During their passage through the wall from the plasma membrane and periplasmic space to the outermost cell wall layers (□, ○) and possibly the extracellular environment (•, ▪), proteins and glycoproteins are most likely to be in equilibrium with other proteinaceous constituents, thus contributing, at least from a functional point of view, to the cell wall layering. In any case, protein and glycoprotein species other than those specifically secreted to the exocellular medium may also be released to such locations by dying (lysed) cells or as a consequence of unbalanced processes of synthesis and degradation of the cell wall structure, required for wall expansion during cell growth. To simplify the scheme, some aspects such as possible interactions of cell wall components with the plasma membrane and proteins retained in the cell wall, apparently by either covalent or noncovalent linkages, are not depicted.
Similar articles
- Initial steps of wall protoplast regeneration in Candida albicans.
Rico H, Carrillo C, Aguado C, Mormeneo S, Sentandreu R. Rico H, et al. Res Microbiol. 1997 Sep-Oct;148(7):593-603. doi: 10.1016/S0923-2508(97)88083-7. Res Microbiol. 1997. PMID: 9765844 - Serologic response to cell wall mannoproteins and proteins of Candida albicans.
Martínez JP, Gil ML, López-Ribot JL, Chaffin WL. Martínez JP, et al. Clin Microbiol Rev. 1998 Jan;11(1):121-41. doi: 10.1128/CMR.11.1.121. Clin Microbiol Rev. 1998. PMID: 9457431 Free PMC article. Review. - Sequential fractionation and two-dimensional gel analysis unravels the complexity of the dimorphic fungus Candida albicans cell wall proteome.
Pitarch A, Sánchez M, Nombela C, Gil C. Pitarch A, et al. Mol Cell Proteomics. 2002 Dec;1(12):967-82. doi: 10.1074/mcp.m200062-mcp200. Mol Cell Proteomics. 2002. PMID: 12543933 - Beta-1,2 oligomannose adhesin epitopes are widely distributed over the different families of Candida albicans cell wall mannoproteins and are associated through both N- and O-glycosylation processes.
Fradin C, Slomianny MC, Mille C, Masset A, Robert R, Sendid B, Ernst JF, Michalski JC, Poulain D. Fradin C, et al. Infect Immun. 2008 Oct;76(10):4509-17. doi: 10.1128/IAI.00368-08. Epub 2008 Jul 21. Infect Immun. 2008. PMID: 18644880 Free PMC article. - Impact of the Environment upon the Candida albicans Cell Wall and Resultant Effects upon Immune Surveillance.
Childers DS, Avelar GM, Bain JM, Larcombe DE, Pradhan A, Budge S, Heaney H, Brown AJP. Childers DS, et al. Curr Top Microbiol Immunol. 2020;425:297-330. doi: 10.1007/82_2019_182. Curr Top Microbiol Immunol. 2020. PMID: 31781866 Review.
Cited by
- Fungal chitinases: diversity, mechanistic properties and biotechnological potential.
Hartl L, Zach S, Seidl-Seiboth V. Hartl L, et al. Appl Microbiol Biotechnol. 2012 Jan;93(2):533-43. doi: 10.1007/s00253-011-3723-3. Epub 2011 Dec 2. Appl Microbiol Biotechnol. 2012. PMID: 22134638 Free PMC article. Review. - Comparison of cell wall proteins in putative Candida albicans & Candida dubliniensis by using modified staining method & SDS-PAGE.
Yazdanparast SA, Nezarati SS, Heshmati F, Hamzehlou S. Yazdanparast SA, et al. Med J Islam Repub Iran. 2012 May;26(2):45-9. Med J Islam Repub Iran. 2012. PMID: 23482280 Free PMC article. - The actin-regulating kinase homologue MoArk1 plays a pleiotropic function in Magnaporthe oryzae.
Wang J, Du Y, Zhang H, Zhou C, Qi Z, Zheng X, Wang P, Zhang Z. Wang J, et al. Mol Plant Pathol. 2013 Jun;14(5):470-82. doi: 10.1111/mpp.12020. Epub 2013 Feb 5. Mol Plant Pathol. 2013. PMID: 23384308 Free PMC article. - Transcriptional Control of Drug Resistance, Virulence and Immune System Evasion in Pathogenic Fungi: A Cross-Species Comparison.
Pais P, Costa C, Cavalheiro M, Romão D, Teixeira MC. Pais P, et al. Front Cell Infect Microbiol. 2016 Oct 20;6:131. doi: 10.3389/fcimb.2016.00131. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 27812511 Free PMC article. Review. - Candida albicans cell wall proteins.
Chaffin WL. Chaffin WL. Microbiol Mol Biol Rev. 2008 Sep;72(3):495-544. doi: 10.1128/MMBR.00032-07. Microbiol Mol Biol Rev. 2008. PMID: 18772287 Free PMC article. Review.
References
- Aguiar J M, Baquero F, Jones J M. Candida albicans exocellular antigens released into a synthetic culture medium: characterization and serological response in rabbits. J Gen Microbiol. 1993;139:3005–3010. - PubMed
- Akashi T, Homma M, Kanbe T, Tanaka K. Ultrastructure of proteinase-secreting cells of Candida albicans studied by alkaline bismuth staining and immunocytochemistry. J Gen Microbiol. 1993;139:2185–2195. - PubMed
- Alloush H M, López-Ribot J L, Chaffin W L. Dynamic expression of cell wall proteins of Candida albicans revealed by probes from cDNA clones. J Med Vet Mycol. 1996;34:91–97. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources