The Saccharomyces cerevisiae SCS2 gene product, a homolog of a synaptobrevin-associated protein, is an integral membrane protein of the endoplasmic reticulum and is required for inositol metabolism - PubMed (original) (raw)
The Saccharomyces cerevisiae SCS2 gene product, a homolog of a synaptobrevin-associated protein, is an integral membrane protein of the endoplasmic reticulum and is required for inositol metabolism
S Kagiwada et al. J Bacteriol. 1998 Apr.
Abstract
The Saccharomyces cerevisiae SCS2 gene has been cloned as a suppressor of inositol auxotrophy of CSE1 and hac1/ire15 mutants (J. Nikawa, A. Murakami, E. Esumi, and K. Hosaka, J. Biochem. 118:39-45, 1995) and has homology with a synaptobrevin/VAMP-associated protein, VAP-33, cloned from Aplysia californica (P. A. Skehel, K. C. Martin, E. R. Kandel, and D. Bartsch, Science 269:1580-1583, 1995). In this study we have characterized an SCS2 gene product (Scs2p). The product has a molecular mass of 35 kDa and is C-terminally anchored to the endoplasmic reticulum, with the bulk of the protein located in the cytosol. The disruption of the SCS2 gene causes yeast cells to exhibit inositol auxotrophy at temperatures of above 34 degrees C. Genetic studies reveal that the overexpression of the INO1 gene rescues the inositol auxotrophy of the SCS2 disruption strain. The significant primary structural feature of Scs2p is that the protein contains the 16-amino-acid sequence conserved in yeast and mammalian cells. The sequence is required for normal Scs2p function, because a mutant Scs2p that lacks the sequence does not complement the inositol auxotrophy of the SCS2 disruption strain. Therefore, the Scs2p function might be conserved among eukaryotic cells.
Figures
FIG. 1
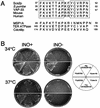
Inositol auxotrophy of _scs2_Δ strains. KY360 (_scs2_Δ::TRP1) cells transformed with YCp_lac_33 (control), YCp(SCS2), or YCp(HA-SCS2) were streaked for isolation on either inositol-containing (INO+) or inositol-free (INO−) minimal medium and incubated at 34°C for 72 h or at 37°C for 96 h.
FIG. 2
Inositol auxotrophy of _scs2_Δ strains transformed with INO1 or INO2. KY360 (_scs2_Δ::TRP1) cells transformed with YCp_lac_33 [vector control for YCp(INO1)], YEplac195 [vector control for YEp(INO2)], YCp(INO1), or YEp(INO2) were streaked for isolation on either inositol-containing (INO+), or inositol-free (INO−) minimal medium and incubated at 34°C for 72 h.
FIG. 3
Characterization of Scs2p by Western blotting. (A) Strains were grown in minimal medium, converted to spheroplasts, and lysed osmotically. The resultant lysate was centrifuged at 800 × g to yield LSS. The LSS was separated by SDS–10% PAGE and immunoblotted with the anti-Scs2p polyclonal antibody (α-Scs2p) (lanes 1 to 3) or the anti-HA monoclonal antibody (α-HA) (lanes 4 and 5). The strains employed were YPH500 (wild type [WT]) (lane 1) and KY360 (_scs2_Δ::TRP1) harboring either YCp_lac_33 (lanes 2 and 4) or YCp(HA-SCS2) (lanes 3 and 5). (B) The LSS fraction of wild-type cells (CTY182) was subjected to two rounds of differential centrifugation to yield 12,000 × g pellet and supernatant fractions (12P and 12S, respectively) and 100,000 × g pellet and supernatant fractions (100P and 100S, respectively). These fractions were resolved by SDS–10% PAGE and immunoblotted for the presence of Scs2p and for markers of the ER membrane (Dpm1p), ER lumen (Kar2p), and cytoplasm/Golgi membrane (Sec14p). (C) The 12P fraction of wild-type cells (CTY182) was incubated with 0.2 volume of one of the following solutions: H2O (control), 5 M KCl, 10 M urea, 0.5 M Na2CO3 (pH 11), or 5% Triton X-100 (TX-100). After incubation at 4°C for 30 min, samples were centrifuged at 100,000 × g for 30 min to separate supernatant (S) and pellet (P) fractions. These fractions were resolved by SDS–10% PAGE and immunoblotted with the anti-Scs2p polyclonal antibody. (D) The 12P fraction of KY360 (_scs2_Δ::TRP1) harboring YCp(HA-SCS2) was incubated with 0.1 mg of trypsin per ml in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of 0.1% Triton X-100 (TX-100). Soybean trypsin inhibitor (STI) was added at the beginning (lanes 3 and 4) or end (lanes 1 and 2) of the incubation. Samples were resolved by SDS–10% PAGE and immunoblotted with the anti-HA monoclonal antibody (top panel) or the anti-Kar2p polyclonal antibody (bottom panel). In panels A and C, the numbers on the left are molecular masses in kilodaltons.
FIG. 4
Localization of Scs2p. Indirect immunofluorescence was carried out with YPH500 cells (wild type) (a, b, and c), KY360 cells (_scs2_Δ::TRP1) (d, e, and f), and KY360 cells harboring YCp(HA-SCS2) (g, h, and i). The cells in panels a to f were incubated with the rabbit anti-Scs2p polyclonal antibody and the mouse anti-Dpm1p monoclonal antibody, and cells in panels g to i were incubated with the rabbit anti-HA polyclonal antibody and the mouse anti-Dpm1p monoclonal antibody. Samples were stained with tetramethylrhodamine isothiocyanate-conjugated anti-rabbit IgG to detect Scs2p (a, d, and g), fluorescein isothiocyanate-conjugated anti-mouse IgG to detect Dpm1p (b, e, and h), and DAPI to indicate the position of the nuclei (c, f, and i).
FIG. 5
The 16-amino-acid conserved region is required for Scs2p activity. (A) Alignment of a 16-amino-acid sequence of Scs2p with those of S. pombe, A. californica (VAP-33), and mouse and human homologs. The sequences of the mouse and human homologs are deduced from nucleotide sequences of expressed sequence tags. Residues identical in all five sequences are boxed. Homologous sequences of MSP1A, TER ATPase, and Cdc48p are also shown. For Scs2p, the S. pombe homolog, VAP-33, MSP1A, TER ATPase, and Cdc48p, the numbers refer to amino acid positions. GenBank accession numbers are D44493 (Scs2p), Z73099 (S. pombe), U36779 (VAP-33), W54842 (mouse), N34715 (human), P53021 (MSP1A), U11760 (TER ATPase), and X56956 (Cdc48p). (B) Overproduction of Scs2pΔ36-53 failed to suppress the inositol auxotrophy of the _scs2_Δ strain. KY360 (_scs2_Δ::TRP1) cells transformed with YEp_lac_195, YEp(SCS2), YEp(_SCS2_Δ36-53), YCp_lac_33, YCp(SCS2), or YCp(_SCS2_Δ36-53) were streaked for isolation on either inositol-containing (INO+) or inositol-free (INO−) minimal medium and incubated at 34°C for 72 h or at 37°C for 96 h.
FIG. 6
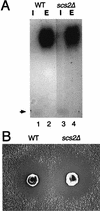
Disruption of SCS2 has no effect on protein secretion. (A) Sugar modification of invertase. Cells were grown in YPD medium at 25°C and shifted to YP medium with 0.1% glucose. After 2 h of incubation at 30°C, cells were converted to spheroplasts and separated into the intracellular (I) and extracellular (E) fractions. These fractions were subjected to native gel activity staining for invertase. The position of nonglycosylated (cytosolic) invertase is indicated by an arrow. Strains used were CTY182 (wild type [WT]) (lanes 1 and 2) and KY356 (_scs2_Δ::URA3) (lanes 3 and 4). (B) Halo assay for α-factor production. YPH500 (WT) and KY360 (_scs2_Δ::TRP1) cells were spotted on a lawn of a-mating-type cells (MATa sst2) on a plate and incubated at 30°C for 48 h to allow halos to develop.
Similar articles
- Cloning and sequence of the SCS2 gene, which can suppress the defect of INO1 expression in an inositol auxotrophic mutant of Saccharomyces cerevisiae.
Nikawa J, Murakami A, Esumi E, Hosaka K. Nikawa J, et al. J Biochem. 1995 Jul;118(1):39-45. doi: 10.1093/oxfordjournals.jbchem.a124889. J Biochem. 1995. PMID: 8537323 - Role of the yeast VAP homolog, Scs2p, in INO1 expression and phospholipid metabolism.
Kagiwada S, Zen R. Kagiwada S, et al. J Biochem. 2003 Apr;133(4):515-22. doi: 10.1093/jb/mvg068. J Biochem. 2003. PMID: 12761300 - The yeast VAP homolog Scs2p has a phosphoinositide-binding ability that is correlated with its activity.
Kagiwada S, Hashimoto M. Kagiwada S, et al. Biochem Biophys Res Commun. 2007 Dec 28;364(4):870-6. doi: 10.1016/j.bbrc.2007.10.079. Epub 2007 Oct 24. Biochem Biophys Res Commun. 2007. PMID: 17963691 - Interaction between repressor Opi1p and ER membrane protein Scs2p facilitates transit of phosphatidic acid from the ER to mitochondria and is essential for INO1 gene expression in the presence of choline.
Gaspar ML, Chang YF, Jesch SA, Aregullin M, Henry SA. Gaspar ML, et al. J Biol Chem. 2017 Nov 10;292(45):18713-18728. doi: 10.1074/jbc.M117.809970. Epub 2017 Sep 18. J Biol Chem. 2017. PMID: 28924045 Free PMC article. - Desumoylation of the endoplasmic reticulum membrane VAP family protein Scs2 by Ulp1 and SUMO regulation of the inositol synthesis pathway.
Felberbaum R, Wilson NR, Cheng D, Peng J, Hochstrasser M. Felberbaum R, et al. Mol Cell Biol. 2012 Jan;32(1):64-75. doi: 10.1128/MCB.05878-11. Epub 2011 Oct 24. Mol Cell Biol. 2012. PMID: 22025676 Free PMC article.
Cited by
- ERG30, a VAP-33-related protein, functions in protein transport mediated by COPI vesicles.
Soussan L, Burakov D, Daniels MP, Toister-Achituv M, Porat A, Yarden Y, Elazar Z. Soussan L, et al. J Cell Biol. 1999 Jul 26;146(2):301-11. doi: 10.1083/jcb.146.2.301. J Cell Biol. 1999. PMID: 10427086 Free PMC article. - Gene recruitment of the activated INO1 locus to the nuclear membrane.
Brickner JH, Walter P. Brickner JH, et al. PLoS Biol. 2004 Nov;2(11):e342. doi: 10.1371/journal.pbio.0020342. Epub 2004 Sep 28. PLoS Biol. 2004. PMID: 15455074 Free PMC article. - The amyotrophic lateral sclerosis 8 protein VAPB is cleaved, secreted, and acts as a ligand for Eph receptors.
Tsuda H, Han SM, Yang Y, Tong C, Lin YQ, Mohan K, Haueter C, Zoghbi A, Harati Y, Kwan J, Miller MA, Bellen HJ. Tsuda H, et al. Cell. 2008 Jun 13;133(6):963-77. doi: 10.1016/j.cell.2008.04.039. Cell. 2008. PMID: 18555774 Free PMC article. - The Interactome of the VAP Family of Proteins: An Overview.
James C, Kehlenbach RH. James C, et al. Cells. 2021 Jul 14;10(7):1780. doi: 10.3390/cells10071780. Cells. 2021. PMID: 34359948 Free PMC article. Review. - Gene encoding vesicle-associated membrane protein-associated protein from Triticum aestivum (TaVAP) confers tolerance to drought stress.
Singh B, Khurana P, Khurana JP, Singh P. Singh B, et al. Cell Stress Chaperones. 2018 May;23(3):411-428. doi: 10.1007/s12192-017-0854-1. Epub 2017 Nov 7. Cell Stress Chaperones. 2018. PMID: 29116579 Free PMC article.
References
- Ambroziak J, Henry S A. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J Biol Chem. 1994;269:15344–15349. - PubMed
- Carman G M, Henry S A. Phospholipid biosynthesis in yeast. Annu Rev Biochem. 1989;58:635–669. - PubMed
- Cox J S, Shamu C E, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous