The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1 - PubMed (original) (raw)
The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1
G Chuck et al. Genes Dev. 1998.
Abstract
The orderly production of meristems with specific fates is crucial for the proper elaboration of plant architecture. The maize inflorescence meristem branches several times to produce lateral meristems with determinate fates. The first meristem formed, the spikelet pair meristem, produces two spikelet meristems, each of which produces two floral meristems. We have identified a gene called indeterminate spikelet1 (ids1) that specifies a determinate spikelet meristem fate and thereby limits the number of floral meristems produced. In the absence of ids1 gene function, the spikelet meristem becomes indeterminate and produces additional florets. Members of the grass family vary in the number of florets within their spikelets, suggesting that ids1 may play a role in inflorescence architecture in other grass species. ids1 is a member of the APETALA2 (AP2) gene family of transcription factors that has been implicated in a wide range of plant development roles. Expression of ids1 was detected in many types of lateral organ primordia as well as spikelet meristems. Our analysis of the ids1 mutant phenotype and expression pattern indicates that ids1 specifies determinate fates by suppressing indeterminate growth within the spikelet meristem.
Figures
Figure 1
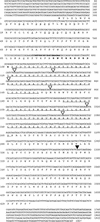
Nucleotide and deduced amino acid sequence of IDS1. Amino acid numbers are shown to the left; nucleotides are numbered on the right. Underlined amino acid sequences represent the two AP2 domains (Jofuku et al. 1994). Serines and acidic amino acids at the amino terminus are indicated with short underlines. Amino acid sequences in bold represent the basic region with putative nuclear localization activity. Bold nucleotide sequences correspond to the ZAP2-1 primer used for the PCR screen. (▿) The six intron positions conserved between IDS, AP2, and GL15; (▾) the other intron.
Figure 2
Comparison between the AP2 domains of the _AP2_-like genes. The deduced amino acid sequences of the AP2 domains of IDS1 (GenBank accession no. AF048900), RAP2.7 (AF003100), GL15 (U41466), and ANT (U40256) are compared with AP2 (ATU12546). The region in between the two AP2 repeats, i.e., the linker region, starts at amino acid 78 and ends at 102. Gaps, represented by dots, were introduced to facilitate the alignment. Amino acids shared by all five proteins are shown in boldface type and on the consensus sequence at bottom.
Figure 3
Localization of the Mu elements in ids1–mum1 and ids1–mum2. ids1 intron and exon sequences are shown in normal and boldface lettering, respectively. Intron and exon numbers are shown above the DNA sequences. The underlined sequences represent the characteristic 9-bp duplication associated with Mu insertions.
Figure 4
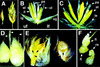
Phenotypes in ids1–mum male and female spikelets. (A) Normal A632 tassel spikelet (left) and ids1–mum2 tassel spikelet (right). (B) Dissected normal A632 tassel spikelet showing lateral organs of upper and lower florets. (C) Dissected ids1–mum2 tassel spikelet showing four florets instead of two. Each floret has three stamens surrounded by palea and lemma as in A632. (D) Normal A632 ear spikelet (left) and ids–mum1 ear spikelet (right). Arrowheads point to silks. (E) Median longitudinal sections through unfertilized 25-day-old ids1–mum2 ear spikelet (left) and A632 ear spikelet (right). Black arrows point to florets. A632 spikelets have a single floret with an enlarged nucellus; ids1–mum2 ear spikelets have at least three florets with greatly reduced lateral organ development. (F) ids1–mum2 ear spikelet with an elongating rachilla containing two florets. The single arrowhead points to the tip of the rachilla. (st) Stamen; (il) inner lemma; (pa) palea; (ol) outer lemma; (ig) inner glume; (og) outer glume; (uf) upper floret; (lf) lower floret; (ra) rachilla; (fl) floret.
Figure 5
Scanning electron microscopy of normal and ids1–mum2 ears. (A) A632 ear primordium with initiating spikelet pair primordia and spikelet primordia. Bar, 300 μm. (B) A632 unbranched spikelet meristem (top) and slightly older branched spikelet meristem (bottom). The older spikelet meristem undergoes lateral branching to initiate the lower floret. Bar, 102 μm. (C) Development of lateral organs in A632 ear spikelets. The lateral organs of the upper floret develop first. Bar, 80 μm. (D) Maturing lateral organs of the upper floret of A632 ear spikelets with initiating gynoecial ridge. Bar, 104 μm. (E) Branching ids1–mum2 spikelet meristems. The uppermost spikelet meristem has not branched yet and appears elongated. Simultaneous floret branching and lemma initiation occurs on opposite sides of the middle and lower spikelet meristems. Bar, 156 μm. (F) Further development of ids1–mum2 spikelet meristems with glumes removed showing development of the floral meristems within the axils of the lemmas. The spikelet meristem persists after each branching event. Bar, 58 μm. (G) Older ids1–mum2 spikelet meristem with glumes removed. The residual spikelet meristem elongates to initiate another floret. Bar, 60 μm. (H) ids1–mum2 spikelet showing maturing lateral organs. There is no distinction between upper and lower floret in terms of lateral organ development. The youngest floret branching from the spikelet meristem is obscured by the lemma. Bar, 171 μm. (spm) Spikelet pair meristem; (sm) spikelet meristem; (lfm) lower floral meristem; (gy) gynoecium; (fm) floral meristem; (le) lemma.
Figure 6
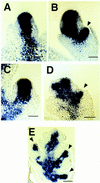
In situ localization of kn1 within A632 and ids1–mum2 spikelet meristems. (A) kn1 expression within an unbranched A632 spikelet meristem. Bar, 50 μm. (B) kn1 expression within a branching A632 spikelet meristem. Expression can be seen in the initiating lower floret. Bar, 50 μm. (C) kn1 expression within an unbranched ids1–mum2 spikelet meristem. Bar, 50 μm. (D) kn1 expression within an early branching ids1–mum2 spikelet meristem. Initiating florets can be seen on both sides of the spikelet meristem. Bar, 60 μm. (E) kn1 expression within an older ids1–mum2 spikelet. Strong expression persists within the developing florets. Bar, 80 μm. Arrowheads point to initiating florets in B, D, and E.
Figure 7
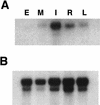
Analysis of ids1 expression in different organs of B73 maize. (A) An RNA gel blot containing 1 μg of poly(A)+ RNA isolated from embryos 17 days postpollination (E), shoot meristems including unexpanded stem and the youngest leaf primordia (M), ear inflorescence primordia up to 2 cm long (I), primary roots from seeds germinated in humid air (R), and the blade portion of fully expanded juvenile leaves (L) was blotted and hybridized to the 3′ portion of the ids1 cDNA that does not include the AP2 domain. (B) Control hybridization. The blot in A was probed with the maize ubiquitin cDNA to show relative loading.
Figure 8
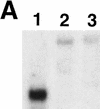
Analysis of ids1 expression in ids1–mum mutant ears. (A) An RNA gel containing 1 μg of poly(A)+ RNA isolated from 2-cm ears from A632 (1), ids1–mum2 (2), and ids1–mum1 (3) was blotted and hybridized to the 3′ portion of the ids1 cDNA. (B) Control hybridization. The blot in A was probed with the maize ubiquitin cDNA to show relative loading.
Figure 8
Analysis of ids1 expression in ids1–mum mutant ears. (A) An RNA gel containing 1 μg of poly(A)+ RNA isolated from 2-cm ears from A632 (1), ids1–mum2 (2), and ids1–mum1 (3) was blotted and hybridized to the 3′ portion of the ids1 cDNA. (B) Control hybridization. The blot in A was probed with the maize ubiquitin cDNA to show relative loading.
Figure 9
In situ localization in B73 vegetative and floral apices by use of the 3′ portion of the ids1 cDNA. (A) Shoot apical meristem with young leaf primordia (P1 and P2) and incipient leaf primordia (P0). Bar, 65 μm. (B) Young ear inflorescence. ids1 expression can be seen in the acropetally initiating spikelet pair primordia. Bar, 70 μm. (C) Unbranched spikelet meristems. Expression can be seen in the spikelet meristem but not in the glumes. Bar, 36 μm. (D) Branching spikelet meristems. Strong expression can be seen in the inner and outer lemmas as well as the zone in between the initiating upper and lower florets (unlabeled arrow). This zone of expression gradually disappears in the older spikelet below. No expression is detected in the initiating floral meristems. Bar, 70 μm. (E) Upper floret with initiating stamens. Discrete areas of ids1 expression can be observed in the initiating stamen primordia and palea. ids1 expression persists in the expanding inner and outer lemmas. Bar, 70 μm. (F) Older floret with lodicules. Stamen expression now localizes to the locule chambers of the anther. No expression can be seen in the lodicules (lo). Bar, 70 μm. (G) ids1–mum2 spikelet probed with ids1. Little or no ids1 transcript can be detected. Arrowheads point to florets. Bar, 130 μm.
Figure 10
Models for maize floret initiation. (A) Terminal upper floret model. The spikelet meristem branches once laterally to form the lower floret. The residual spikelet meristem (shown in black) is then transformed into the upper floret (black). (B) Lateral branching model. The spikelet meristem undergoes lateral branching to form the lower and then upper florets. The residual spikelet meristem (shown in black) is found in a small region in between the florets. A rudiment of the spikelet meristem in found in the rachilla in the mature spikelet (black).
Similar articles
- Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1.
Chuck G, Meeley R, Hake S. Chuck G, et al. Development. 2008 Sep;135(18):3013-9. doi: 10.1242/dev.024273. Epub 2008 Aug 13. Development. 2008. PMID: 18701544 - The indeterminate floral apex1 gene regulates meristem determinacy and identity in the maize inflorescence.
Laudencia-Chingcuanco D, Hake S. Laudencia-Chingcuanco D, et al. Development. 2002 Jun;129(11):2629-38. doi: 10.1242/dev.129.11.2629. Development. 2002. PMID: 12015291 - Combinatorial control of meristem identity in maize inflorescences.
Kaplinsky NJ, Freeling M. Kaplinsky NJ, et al. Development. 2003 Mar;130(6):1149-58. doi: 10.1242/dev.00336. Development. 2003. PMID: 12571106 - Grass meristems II: inflorescence architecture, flower development and meristem fate.
Tanaka W, Pautler M, Jackson D, Hirano HY. Tanaka W, et al. Plant Cell Physiol. 2013 Mar;54(3):313-24. doi: 10.1093/pcp/pct016. Epub 2013 Jan 31. Plant Cell Physiol. 2013. PMID: 23378448 Review. - Flowering and determinacy in maize.
Bortiri E, Hake S. Bortiri E, et al. J Exp Bot. 2007;58(5):909-16. doi: 10.1093/jxb/erm015. Epub 2007 Mar 3. J Exp Bot. 2007. PMID: 17337752 Review.
Cited by
- A spatial transcriptome map of the developing maize ear.
Wang Y, Luo Y, Guo X, Li Y, Yan J, Shao W, Wei W, Wei X, Yang T, Chen J, Chen L, Ding Q, Bai M, Zhuo L, Li L, Jackson D, Zhang Z, Xu X, Yan J, Liu H, Liu L, Yang N. Wang Y, et al. Nat Plants. 2024 May;10(5):815-827. doi: 10.1038/s41477-024-01683-2. Epub 2024 May 14. Nat Plants. 2024. PMID: 38745100 - Flower development and a functional analysis of related genes in Impatiens uliginosa.
He H, Chen X, Wang T, Zhang X, Liu Z, Qu S, Gu Z, Huang M, Huang H. He H, et al. Front Plant Sci. 2024 Mar 25;15:1370949. doi: 10.3389/fpls.2024.1370949. eCollection 2024. Front Plant Sci. 2024. PMID: 38590746 Free PMC article. - Computational analysis of the AP2/ERF family in crops genome.
Choudhury S. Choudhury S. BMC Genomics. 2024 Jan 23;25(1):102. doi: 10.1186/s12864-024-09970-0. BMC Genomics. 2024. PMID: 38262942 Free PMC article. - FRIZZLE PANICLE (FZP) regulates rice spikelets development through modulating cytokinin metabolism.
Wang W, Chen W, Wang J. Wang W, et al. BMC Plant Biol. 2023 Dec 16;23(1):650. doi: 10.1186/s12870-023-04671-4. BMC Plant Biol. 2023. PMID: 38102566 Free PMC article. - Identification of QTLs and their candidate genes for the number of maize tassel branches in F2 from two higher generation sister lines using QTL mapping and RNA-seq analysis.
Ruidong S, Shijin H, Yuwei Q, Yimeng L, Xiaohang Z, Ying L, Xihang L, Mingyang D, Xiangling L, Fenghai L. Ruidong S, et al. Front Plant Sci. 2023 Aug 13;14:1202755. doi: 10.3389/fpls.2023.1202755. eCollection 2023. Front Plant Sci. 2023. PMID: 37641589 Free PMC article.
References
- Bennetzen JL, Springer PS, Cresse AD, Hendrickx M. Specificity and regulation of the Mutator transposable element system in maize. Critical Rev Plant Sci. 1993;12:57–95.
- Bonnett, O.T. 1953. Developmental morphology of the vegetative and floral shoots of maize. Ill. Agric. Exp. Stn. Bull. 568.
- Bossinger G, Rohde W, Lundqvist U, Salamini F. Genetics of barley development: Mutant phenotypes and molecular aspects, In: Shewry PR, editor. Barley: Genetics, biochemistry, molecular biology and biotechnology. Oxford, UK: Alden Press Ltd; 1992. pp. 231–264.
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development. 1993;119:721–743.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials