Human immunodeficiency virus type 1 attachment to HeLa CD4 cells is CD4 independent and gp120 dependent and requires cell surface heparans - PubMed (original) (raw)
Human immunodeficiency virus type 1 attachment to HeLa CD4 cells is CD4 independent and gp120 dependent and requires cell surface heparans
I Mondor et al. J Virol. 1998 May.
Abstract
The binding of human immunodeficiency virus type 1 (HIV-1) (Hx10) virions to two different cell lines was analyzed by using a novel assay based on the detection, by anti-HLA-DR-specific antibodies, of HLA-DR+ virus binding to HLA-DR- cells. Virion attachment to the CD4+-T-cell line A3.01 was highly CD4 dependent in that it was potently inhibited by CD4 monoclonal antibodies (MAbs), and little virus binding to the CD4- sister A2.01 line was observed. By contrast, virion binding to HeLa cells expressing moderate or high levels of CD4 was equivalent to, or lower than, binding to wild-type CD4- HeLa cells. Moreover, several CD4 MAbs did not reduce, but enhanced, HIV-1 attachment to HeLa-CD4 cells. CD4 was required for infection of HeLa cells, however, demonstrating a postattachment role for this receptor. MAbs specific for the V2 and V3 loops and the CD4i epitope of gp120 strongly inhibited virion binding to HeLa-CD4 cells, whereas MAbs specific for the CD4bs and the 2G12 epitopes enhanced attachment. Despite this, all gp120- and gp41-specific MAbs tested neutralized infectivity on HeLa-CD4 cells. HIV-1 attachment to HeLa cells was only partially inhibited by MAbs specific for adhesion molecules present on the virus or target cells but was completely blocked by polyanions such as heparin, dextran sulfate, and pentosan sulfate. Treatment of HeLa-CD4 cells with heparinases completely eliminated HIV attachment and infection, strongly implicating cell surface heparans in the attachment process. CD4 dependence for HIV-1 attachment to target cells is thus highly cell line specific and may be replaced by other ligand-receptor interactions.
Figures
FIG. 1
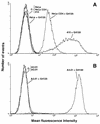
Measurement of CD4 levels on the cell lines used. (A) HeLa, HeLa-CD4, and HeLa-CD4 clone 15 cells; (B) A3.01 and A2.01 cells. Cells in suspension were labeled or not with the CD4 MAb Q4120 for 1 h at 4°C and then washed and stained with anti-mouse IgG–phycoerythrin. Peaks marked C− represent the background staining in the absence of anti-CD4 MAb or CD4− cell lines (A2.01 and wild-type HeLa cells) in the presence of anti-CD4 MAb. Fluorescence was analyzed by flow cytometry; 104 gated events were acquired for each datum point, and these are expressed as the MFI.
FIG. 2
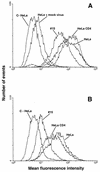
Hx10 attachment to HeLa, HeLa-CD4, and HeLa-CD4 clone 15 cells. Cells in suspension were incubated with undiluted concentrated virus or mock virus for 30 min at 37°C, washed, labeled with biotinylated anti-HLA-DR MAb (A) or gp120 CD4bs-specific MAb IgG1b12 (B), and fixed overnight. The cells were then stained with the appropriate phycoerythrin conjugate, washed, and analyzed by flow cytometry as described for Fig. 1. C-HeLa represents HeLa cells incubated with the phycoerythrin conjugate alone; HeLa-CD4 and HeLa-CD4 clone 15 cells yielded very similar background signals (not shown).
FIG. 3
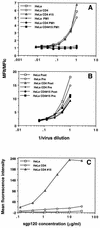
Concentration dependence of Hx10 and sgp120IIIB binding to HeLa, HeLa-CD4, and HeLa-CD4 clone 15 cells. HeLa, HeLa-CD4, and HeLa-CD4 clone 15 cells were incubated with serial dilutions of concentrated virus, mock virus, sgp120 and stained and analyzed as described for Fig. 1. (A) Detection of prebound virus with the biotinylated anti-HLA-DR MAb. Open symbols, incubation with virus; closed sympols, incubation with mock virus. (B) Another experiment in which virus was labeled with anti-HLA-DR MAb prior to cell binding (Pre) or subsequent to cell binding (Post). Results are expressed as the ratio of the test signal to the background signal (MFIt/MFIc) to normalize for variation between the different cell clones in the background staining. (C) HeLa, HeLa-CD4, and HeLa-CD4 clone 15 cells were incubated with sgp120 for 4 h at 4°C before being labeled with anti-gp120, washed, and stained with fluorescein conjugate. Binding was analyzed by flow cytometry as described for Fig. 1.
FIG. 4
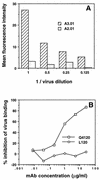
Concentration dependence of Hx10 binding to A3.01 and A2.01 cells and inhibition by CD4 MAbs. (A) A3.01 and A2.01 cells were incubated with serial dilutions of Hx10 for 30 min at 37°C before being washed, labeled with biotinylated anti-HLA-DR, washed, and fixing in formaldehyde. After being stained with streptavidin-phycoerythrin, cells were analyzed by flow cytometry as described for Fig. 1. (B) A3.01 cells were preincubated with CD4 MAbs for 1 h at 4°C before addition of virus. Subsequent steps were as described for panel A.
FIG. 5
(A) HeLa-CD4 cells were preincubated with CD4 MAbs for 1 h at 4°C before treatment with Hx10 for 30 min at 37°C. Cells were washed, labeled with anti-HLA-DR, fixed overnight, and subsequently stained with streptavidin-phycoerythrin and analyzed as described for Fig. 1. Results are means and standard deviations for triplicates, expressed as percent inhibition of virus binding. (B) HeLa-CD4 cells pretreated with CD4 MAbs and incubated with virus as described for panel A were cultured for 36 h before lysis and addition of substrate to detect activation of β-galactosidase activity. The color change was read as OD550, and results are expressed as percent inhibition of HIV infection.
FIG. 6
(A and B) HIV was pretreated with neutralizing anti-Env MAbs at 10 μg/ml or with sCD4 at 3 μg/ml for 1 h at 37°C before incubation with HeLa-CD4 (A) or A3.01 (B) cells for 30 min. After washing, bound virus was detected with anti-HLA-DR MAb as described for Fig. 2A. (C and D) HIV was pretreated with neutralizing anti-Env MAbs at 10 μg/ml or with sCD4 at 3 μg/ml for 1 h at 37°C before incubation with HeLa-CD4 (C) or A3.01 (D) cells for 2 h at 37°C. After culture for 36 h, HeLa-CD4 cells were lysed and analyzed for β-galactosidase activity as described for Fig. 5B, and supernatants from A3.01 cultures were analyzed for cell-free p24 protein by enzyme-linked immunosorbent assay as described in Materials and Methods. Results for both cell types are means and standard deviations for triplicates, expressed as percent inhibition of virus attachment (A and B) and of HIV infection (C and D).
FIG. 7
Inhibition of Hx10 binding to HeLa and HeLa-CD4 cells and infection of HeLa-CD4 cells by polyanions, a polycation, and heparinases. (A and B) Hx10 was preincubated with polyanions and control molecules at 10 μg/ml for 1 h at 37°C before addition of HeLa or HeLa-CD4 cells (A), or HeLa and HeLa-CD4 cells were preincubated with poly-
l
-lysine (10 μg/ml) or heparinases (10 U/ml) for 1 h at 37°C before washing and treatment with virus for 30 min at 37°C (B). Virus binding was detected by biotinylated anti-HLA-DR MAb as described for Fig. 2A. (C and D) Virus was preincubated with polyanions or control molecules for 1 h at 37°C before addition of HeLa-CD4 cells (C), or HeLa-CD4 cells were preincubated with poly-
l
-lysine or heparinases before washing and addition of virus (D). Cells were cultured for 36 h at 37°C before detection of β-galactosidase activity as described for Fig. 5. Results are means and standard deviations for triplicates, expressed as percent inhibition of virus attachment (A and B) and of HIV infection (C and D).
Similar articles
- Selective interactions of polyanions with basic surfaces on human immunodeficiency virus type 1 gp120.
Moulard M, Lortat-Jacob H, Mondor I, Roca G, Wyatt R, Sodroski J, Zhao L, Olson W, Kwong PD, Sattentau QJ. Moulard M, et al. J Virol. 2000 Feb;74(4):1948-60. doi: 10.1128/jvi.74.4.1948-1960.2000. J Virol. 2000. PMID: 10644368 Free PMC article. - Neutralizing antibodies against the V3 loop of human immunodeficiency virus type 1 gp120 block the CD4-dependent and -independent binding of virus to cells.
Valenzuela A, Blanco J, Krust B, Franco R, Hovanessian AG. Valenzuela A, et al. J Virol. 1997 Nov;71(11):8289-98. doi: 10.1128/JVI.71.11.8289-8298.1997. J Virol. 1997. PMID: 9343181 Free PMC article. - Cyanovirin-N binds to gp120 to interfere with CD4-dependent human immunodeficiency virus type 1 virion binding, fusion, and infectivity but does not affect the CD4 binding site on gp120 or soluble CD4-induced conformational changes in gp120.
Esser MT, Mori T, Mondor I, Sattentau QJ, Dey B, Berger EA, Boyd MR, Lifson JD. Esser MT, et al. J Virol. 1999 May;73(5):4360-71. doi: 10.1128/JVI.73.5.4360-4371.1999. J Virol. 1999. PMID: 10196334 Free PMC article. - The Genesis and Future Prospects of Small Molecule HIV-1 Attachment Inhibitors.
Wang T, Kadow JF, Meanwell NA, Krystal M. Wang T, et al. Adv Exp Med Biol. 2022;1366:45-64. doi: 10.1007/978-981-16-8702-0_4. Adv Exp Med Biol. 2022. PMID: 35412134 Review. - Fostemsavir.
[No authors listed] [No authors listed] 2023 Jun 20. LiverTox: Clinical and Research Information on Drug-Induced Liver Injury [Internet]. Bethesda (MD): National Institute of Diabetes and Digestive and Kidney Diseases; 2012–. 2023 Jun 20. LiverTox: Clinical and Research Information on Drug-Induced Liver Injury [Internet]. Bethesda (MD): National Institute of Diabetes and Digestive and Kidney Diseases; 2012–. PMID: 37384748 Free Books & Documents. Review.
Cited by
- Regulation of CCR5 expression in human placenta: insights from a study of mother-to-child transmission of HIV in Malawi.
Joubert BR, Franceschini N, Mwapasa V, North KE, Meshnick SR. Joubert BR, et al. PLoS One. 2010 Feb 15;5(2):e9212. doi: 10.1371/journal.pone.0009212. PLoS One. 2010. PMID: 20169157 Free PMC article. - Selective interactions of polyanions with basic surfaces on human immunodeficiency virus type 1 gp120.
Moulard M, Lortat-Jacob H, Mondor I, Roca G, Wyatt R, Sodroski J, Zhao L, Olson W, Kwong PD, Sattentau QJ. Moulard M, et al. J Virol. 2000 Feb;74(4):1948-60. doi: 10.1128/jvi.74.4.1948-1960.2000. J Virol. 2000. PMID: 10644368 Free PMC article. - Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus.
Kwong PD, Wyatt R, Sattentau QJ, Sodroski J, Hendrickson WA. Kwong PD, et al. J Virol. 2000 Feb;74(4):1961-72. doi: 10.1128/jvi.74.4.1961-1972.2000. J Virol. 2000. PMID: 10644369 Free PMC article. - Syndecan-3 is a dendritic cell-specific attachment receptor for HIV-1.
de Witte L, Bobardt M, Chatterji U, Degeest G, David G, Geijtenbeek TB, Gallay P. de Witte L, et al. Proc Natl Acad Sci U S A. 2007 Dec 4;104(49):19464-9. doi: 10.1073/pnas.0703747104. Epub 2007 Nov 26. Proc Natl Acad Sci U S A. 2007. PMID: 18040049 Free PMC article. - Syndecan-Fc hybrid molecule as a potent in vitro microbicidal anti-HIV-1 agent.
Bobardt MD, Chatterji U, Schaffer L, de Witte L, Gallay PA. Bobardt MD, et al. Antimicrob Agents Chemother. 2010 Jul;54(7):2753-66. doi: 10.1128/AAC.01606-09. Epub 2010 May 3. Antimicrob Agents Chemother. 2010. PMID: 20439611 Free PMC article.
References
- Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a rantes, MIP-1a, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. - PubMed
- Armstrong S J, McInerney T L, McLain L, Wahren B, Hinkula J, Levi M, Dimmock N J. Two neutralizing anti-V3 monoclonal antibodies act by affecting different functions of human immunodeficiency virus type 1. J Gen Virol. 1996;77:2931–2941. - PubMed
- Arthur L O, Bess J W, Jr, de Sowder R C, Benveniste R E, Mann D L, Chermann J C, Henderson L E. Cellular proteins bound to immunodeficiency viruses: implications for pathogenesis and vaccines. Science. 1992;258:1935–1938. - PubMed
- Barbas C F I, Collett T A, Amberg W, Roben P, Binley D, Hoekstra D, Cababa D, Jones T M, Williamson R A, Pilkington G R, Haigwood N L, Satterthwait A C, Sanz I, Burton D R. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J Mol Biol. 1993;230:812–823. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials