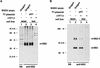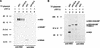Bovine viral diarrhea virus strain Oregon: a novel mechanism for processing of NS2-3 based on point mutations - PubMed (original) (raw)
Bovine viral diarrhea virus strain Oregon: a novel mechanism for processing of NS2-3 based on point mutations
B M Kümmerer et al. J Virol. 1998 May.
Abstract
Bovine viral diarrhea virus (BVDV) isolates can either be cytopathogenic (cp) or noncytopathogenic (noncp). While both biotypes express the nonstructural protein NS2-3, generation of NS3 strictly correlates with the cp phenotype. The production of NS3 is usually caused by cp specific genome alterations, which were found to be due to RNA recombination. Molecular analyses of the cp BVDV strain Oregon revealed that it does not possess such genome alterations but nevertheless is able to generate NS3 via processing of NS2-3. The NS3 serine protease is not involved in this cleavage, which, according to protein sequencing, occurs between amino acids 1589 and 1590 of the BVDV Oregon polyprotein. Transient-expression studies indicated that important information for the cleavage of NS2-3 is located within NS2. This was verified by expression of chimeric constructs containing cDNA fragments derived from BVDV Oregon and a noncp BVDV. It could be shown that the C-terminal part of NS2 plays a crucial role in NS2-3 cleavage. These data, together with results obtained by site-specific exchanges in this region, revealed a new mechanism for NS2-3 processing which is based on point mutations within NS2.
Figures
FIG. 1
Schematic representation of the cDNA clones derived from the BVDV Oregon genome. The upper part shows a scheme of the BVDV genome together with the encoded polyprotein. The structural proteins are shown as shaded boxes, and the nonstructural proteins are shown as open boxes. Below, the cDNA clones are shown. The position and size of the clones obtained after cDNA library screening (A) and RT-PCR (B) are given. The indicated numbers represent the first and last nucleotides of each cDNA clone and refer to the sequence of BVDV SD-1 (11).
FIG. 2
Schematic drawing of the constructs used for transient expression in eucaryotic cells. The diagram at the top represents the BVDV polyprotein, with the structural proteins shown as shaded boxes and the nonstructural proteins shown as open boxes. Below, the expression constructs are presented as lines and drawn to scale to indicate the region of the Oregon polyprotein expressed. The names of the constructs are given on the left. The numbers on the right indicate the amino acids of the BVDV Oregon polyprotein expressed by each construct. Numbers refer to the sequence of BVDV Oregon. In pO1/S-A, inactivation of the NS3 proteinase by a serine-to-alanine change at position 1752 is indicated.
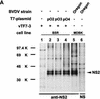
FIG. 3
SDS-PAGE analysis of immunoprecipitates obtained after transient expression of pO1 in BSR cells. For production of authentic BVDV proteins, MDBK cells were infected with BVDV Oregon. Metabolic labeling was performed with [35S]methionine-[35S]cysteine. Nontransfected BSR cells infected with T7 vaccinia virus (vTF7-3) were used as a control. BVDV-specific proteins are indicated on the right. Numbers on the left refer to the molecular masses (in kilodaltons [K]) of marker proteins. +, presence; −, absence. (A) The anti-NS2 serum, which is directed against the carboxy-terminal part of NS2, was used to precipitate NS2 (54). A rabbit preimmune serum (NS) served as a control. (B) Precipitation was carried out with an anti-NS3 serum (56), which recognizes NS2-3 and NS3, or with a rabbit preimmune serum (NS).
FIG. 4
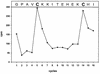
N-terminal sequencing of NS3 from BVDV Oregon. NS3 was generated by transient expression of pO1 in cells metabolically labeled with [35S]cysteine. The graph shows the distribution of radioactivity in counts per minute released during automated Edman degradation after subtraction of background radiation. At the top of the diagram, the amino acid sequence of BVDV Oregon beginning with glycine 1590 is aligned with the degradation steps. The labeled residues are shown in boldface type.
FIG. 5
SDS-PAGE analysis after transient expression of pO1 and pO1/S-A in BSR cells metabolically labeled with [35S]methionine-[35S]cysteine. BVDV-specific proteins are indicated on the right. The truncated NS4B protein is marked with an asterisk. For further details, see the legend to Fig. 3. (A) Precipitation was carried out with the anti-NS2 or anti-NS4 serum. (B) Sera directed against NS3 or NS4 were used for precipitation. NS2-3/4A/4B* and NS3/4A/4B* represent fusion proteins composed of the respective proteins.
FIG. 6
SDS-PAGE of immunoprecipitates after transient expression of the indicated plasmids in BSR cells metabolically labeled with [35S]methionine-[35S]cysteine. For details, see the legend to Fig. 3. (A) Precipitation was carried out with the anti-NS2 serum or a rabbit preimmune serum (NS). (B) The anti-NS2 serum or a preimmune serum (NS) were used for precipitation. NS3# represents an aberrant cleavage product migrating slightly slower than NS3. NS3# is detectable after precipitation with the anti-NS2 serum as well as after precipitation with the anti-NS3 serum (see Fig. 6C). The arrows in lanes 4, 5, and 6 mark the N-terminally truncated NS2-3 proteins. (C) Precipitation with a serum directed against NS3 or a rabbit preimmune serum (NS). See also the legend to panel B.
FIG. 6
SDS-PAGE of immunoprecipitates after transient expression of the indicated plasmids in BSR cells metabolically labeled with [35S]methionine-[35S]cysteine. For details, see the legend to Fig. 3. (A) Precipitation was carried out with the anti-NS2 serum or a rabbit preimmune serum (NS). (B) The anti-NS2 serum or a preimmune serum (NS) were used for precipitation. NS3# represents an aberrant cleavage product migrating slightly slower than NS3. NS3# is detectable after precipitation with the anti-NS2 serum as well as after precipitation with the anti-NS3 serum (see Fig. 6C). The arrows in lanes 4, 5, and 6 mark the N-terminally truncated NS2-3 proteins. (C) Precipitation with a serum directed against NS3 or a rabbit preimmune serum (NS). See also the legend to panel B.
FIG. 6
SDS-PAGE of immunoprecipitates after transient expression of the indicated plasmids in BSR cells metabolically labeled with [35S]methionine-[35S]cysteine. For details, see the legend to Fig. 3. (A) Precipitation was carried out with the anti-NS2 serum or a rabbit preimmune serum (NS). (B) The anti-NS2 serum or a preimmune serum (NS) were used for precipitation. NS3# represents an aberrant cleavage product migrating slightly slower than NS3. NS3# is detectable after precipitation with the anti-NS2 serum as well as after precipitation with the anti-NS3 serum (see Fig. 6C). The arrows in lanes 4, 5, and 6 mark the N-terminally truncated NS2-3 proteins. (C) Precipitation with a serum directed against NS3 or a rabbit preimmune serum (NS). See also the legend to panel B.
FIG. 7
Schematic drawing of the constructs obtained after exchanges of cDNA fragments or single codons in the NS2 coding region. All constructs encode proteins that start within the hypothesized C-terminal part of p7 and end within NS4B. Open boxes indicate sequences derived from BVDV Oregon, whereas shaded boxes represent the CP7Ins− sequence. The CP7Ins− sequence is derived from BVDV CP7 by deletion of the 27-nucleotide insertion within the NS2 gene (54). Chimeric constructs based on plasmid pO1 are shown on the left; chimeric constructs based on pN1 are shown on the right. In each case, the name of the expression construct is given on the left, whereas numbers on the right indicate the positions of the amino acids encoded by the exchanged cDNA fragment or of the exchanged codons, respectively. The abbreviation INS represents the 27-nucleotide insertion specific for BVDV CP7. NS4B*, truncated NS4B.
FIG. 8
Results of transient expression of constructs with heterologous cDNA fragments or point mutations. (A) The upper part shows the SDS-PAGE analysis after transient expression of the chimeric plasmids based on pO1. For details, see the legend to Fig. 3. On top, precipitation was carried out with the anti-NS3 serum. Below, the serum directed against NS2 was used for precipitation. Below the gels, the quantification of the NS2-3 cleavage efficiencies is represented schematically. The quantification was carried out with a phosphorimager after precipitation of NS2-3 and NS3 and SDS-PAGE analysis. For evaluation of cleavage efficiencies, the number of methionine and cysteine residues within each NS2-3 or NS3 protein was determined. After measurement of the radioactivity of the NS2-3 protein, the percentage of the counts resulting from the NS3 moiety was calculated. This value, together with the counts determined for the cleaved NS3 protein, was defined as total NS3 (100%). Cleavage efficiency is equivalent to the percentage of cleaved NS3 with respect to total NS3. Cleavage efficiencies are given as the average of three independent experiments. Values in parentheses are not due to NS3, since in these cases no NS2 could be detected. The exposure time was different for the gels showing NS2-3/NS3 on the one hand and NS2 on the other hand. (B) Results obtained after transient expression of the chimeric plasmids based on pN1. For further details, see the legend to panel A.
FIG. 9
Comparison of sequences encompassing the C-terminal one-third of NS2 from different pestiviruses. Sequences of several cp BVDV strains (Oregon, CP7, NADL, and Osloss), a noncp BVDV strain (SD-1), and a CSFV strain (Alfort-Tübingen) were aligned. Only differences from the BVDV Oregon sequence are specified. Numbers indicate the corresponding position in the BVDV Oregon sequence. The positions of the cIns insertion of BVDV NADL and the ubiquitin (UBI) insertion of BVDV Osloss are shown by vertical lines between adjacent amino acids. Arrows mark the positions of amino acids that have been exchanged between BVDV Oregon and BVDV CP7. The pestivirus sequences are from the following sources: CP7 (35), SD-1 (11), NADL (5), Osloss (10), Alfort-Tübingen (30).
Similar articles
- CRISPR/Cas9-Mediated Knockout of DNAJC14 Verifies This Chaperone as a Pivotal Host Factor for RNA Replication of Pestiviruses.
Isken O, Postel A, Bruhn B, Lattwein E, Becher P, Tautz N. Isken O, et al. J Virol. 2019 Feb 19;93(5):e01714-18. doi: 10.1128/JVI.01714-18. Print 2019 Mar 1. J Virol. 2019. PMID: 30518653 Free PMC article. - Temporal modulation of an autoprotease is crucial for replication and pathogenicity of an RNA virus.
Lackner T, Müller A, Pankraz A, Becher P, Thiel HJ, Gorbalenya AE, Tautz N. Lackner T, et al. J Virol. 2004 Oct;78(19):10765-75. doi: 10.1128/JVI.78.19.10765-10775.2004. J Virol. 2004. PMID: 15367643 Free PMC article. - Pathogenesis of mucosal disease, a deadly disease of cattle caused by a pestivirus.
Tautz N, Meyers G, Thiel HJ. Tautz N, et al. Clin Diagn Virol. 1998 Jul 15;10(2-3):121-7. doi: 10.1016/s0928-0197(98)00037-3. Clin Diagn Virol. 1998. PMID: 9741637 Review. - Non-structural proteins of bovine viral diarrhea virus.
Chi S, Chen S, Jia W, He Y, Ren L, Wang X. Chi S, et al. Virus Genes. 2022 Dec;58(6):491-500. doi: 10.1007/s11262-022-01914-8. Epub 2022 May 25. Virus Genes. 2022. PMID: 35614328 Free PMC article. Review.
Cited by
- Host Cell Receptors Implicated in the Cellular Tropism of BVDV.
Qi S, Wo L, Sun C, Zhang J, Pang Q, Yin X. Qi S, et al. Viruses. 2022 Oct 20;14(10):2302. doi: 10.3390/v14102302. Viruses. 2022. PMID: 36298858 Free PMC article. Review. - CRISPR/Cas9-Mediated Knockout of DNAJC14 Verifies This Chaperone as a Pivotal Host Factor for RNA Replication of Pestiviruses.
Isken O, Postel A, Bruhn B, Lattwein E, Becher P, Tautz N. Isken O, et al. J Virol. 2019 Feb 19;93(5):e01714-18. doi: 10.1128/JVI.01714-18. Print 2019 Mar 1. J Virol. 2019. PMID: 30518653 Free PMC article. - Proposed revision to the taxonomy of the genus Pestivirus, family Flaviviridae.
Smith DB, Meyers G, Bukh J, Gould EA, Monath T, Scott Muerhoff A, Pletnev A, Rico-Hesse R, Stapleton JT, Simmonds P, Becher P. Smith DB, et al. J Gen Virol. 2017 Aug;98(8):2106-2112. doi: 10.1099/jgv.0.000873. Epub 2017 Aug 8. J Gen Virol. 2017. PMID: 28786787 Free PMC article. - Clinical appearance and pathology of cattle persistently infected with bovine viral diarrhoea virus of different genetic subgroups.
Bachofen C, Braun U, Hilbe M, Ehrensperger F, Stalder H, Peterhans E. Bachofen C, et al. Vet Microbiol. 2010 Mar 24;141(3-4):258-67. doi: 10.1016/j.vetmic.2009.09.022. Epub 2009 Sep 25. Vet Microbiol. 2010. PMID: 19819088 Free PMC article.
References
- Baker J C. Bovine viral diarrhea virus: a review. J Am Vet Med Assoc. 1987;190:1449–1458. - PubMed
- Bazan J F, Fletterick R J. Detection of a trypsin-like serine protease domain in flaviviruses and pestiviruses. Virology. 1989;171:637–639. - PubMed
- Becher P, Shannon A D, Tautz N, Thiel H-J. Molecular characterization of border disease virus, a pestivirus from sheep. Virology. 1994;198:542–551. - PubMed
- Collett M S, Larson R, Gold C, Strick D, Anderson D K, Purchio A F. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhea virus. Virology. 1988;165:191–199. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous