fus-1, a pH shift mutant of Semliki Forest virus, acts by altering spike subunit interactions via a mutation in the E2 subunit - PubMed (original) (raw)
fus-1, a pH shift mutant of Semliki Forest virus, acts by altering spike subunit interactions via a mutation in the E2 subunit
S Glomb-Reinmund et al. J Virol. 1998 May.
Abstract
Semliki Forest virus (SFV), an enveloped alphavirus, is a well-characterized paradigm for viruses that infect cells via endocytic uptake and low-pH-triggered fusion. The SFV spike protein is composed of a dimer of E1 and E2 transmembrane subunits, which dissociate upon exposure to low pH, liberating E2 and the fusogenic E1 subunit to undergo independent conformational changes. SFV fusion and infection are blocked by agents such as ammonium chloride, which act by raising the pH in the endosome and inhibiting the low-pH-induced conformational changes in the SFV spike protein. We have previously isolated an SFV mutant, fus-1, that requires more acidic pH to trigger its fusion activity and is therefore more sensitive to inhibition by ammonium chloride. The acid shift in the fusion activity of fus-1 was here shown to be due to a more acidic pH threshold for the initial dissociation of the fus-1 spike dimer, thereby resulting in a more acidic pH requirement for the subsequent conformational changes in both fus-1 E1 and fus-1 E2. Sequence analysis demonstrated that the fus-1 phenotype was due to a mutation in the E2 spike subunit, threonine 12 to isoleucine. fus-1 revertants that have regained the parental fusion phenotype and ammonium chloride sensitivity were shown to have also regained E2 threonine 12. Our results identify a region of the SFV E2 spike protein subunit that regulates the pH dependence of E1-catalyzed fusion by controlling the dissociation of the E1/E2 dimer.
Figures
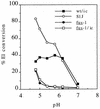
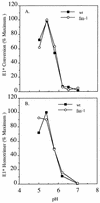
FIG. 1
pH dependence of E1 epitope exposure for wt and mutant viruses. 35S-labeled wt/ic, S1J, fus-1, and fus-1/ic virus preparations were treated at the indicated pH for 10 min at 37°C, adjusted to neutral pH, solubilized in lysis buffer containing 1% Triton X-100, and immunoprecipitated with MAb E1a-1, an acid conformation-specific antibody. E1 precipitated by the MAb was quantitated by SDS-PAGE and phosphorimaging and then compared to the total E1 precipitated by a rabbit polyclonal antibody. Data represent the mean of three separate experiments for each virus. The standard deviations ranged from 1 to 30%.
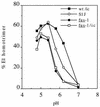
FIG. 2
pH dependence of wt and mutant E1 homotrimer formation. 35S-labeled wt/ic, S1J, fus-1, and fus-1/ic virus preparations were treated at the indicated pH for 10 min at 37°C in the presence of liposomes, adjusted to neutral pH, and solubilized in SDS sample buffer for 3 min at 30°C. Samples were analyzed by SDS-PAGE and quantitated by phosphorimaging. Results are shown as percent E1 in the homotrimer band compared to total E1 and represent the mean for three experiments, with standard deviations between 1 and 33%.
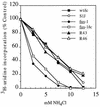
FIG. 3
Sensitivity of infection by wt, mutant, and revertant viruses to inhibition by NH4Cl. BHK cells were pretreated with the indicated concentrations of NH4Cl for 15 min at 37°C and then infected with wt/ic, S1J, fus-1, fus-1/ic, R43 revertant, or R46 revertant at 1 PFU/cell for 1.5 h in the continued presence of NH4Cl. Following infection, the cells were treated with 2 μg of ACD per ml and 15 mM NH4Cl for 30 min and then labeled with [3H]uridine for 3.5 h in the continued presence of ACD and 15 mM NH4Cl. Infection was quantitated as the percent [3H]uridine incorporation compared to controls infected in the absence of NH4Cl. Background incorporation by uninfected cells was subtracted from all points. Data represent the mean of three separate experiments for each virus, with standard deviations between 1 and 20%.
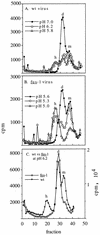
FIG. 4
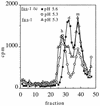
Sucrose gradient sedimentation profiles of wt and fus-1 viral spike proteins. 35S-labeled wt (A and C) or fus-1 (B and C) virus was mixed with 1 mM liposomes, treated at the indicated pH for 10 min at 37°C, solubilized in 1.0 to 0.2% NP-40, and adjusted to pH 7.0. The samples were analyzed by centrifugation on 5 to 20% (wt/wt) sucrose gradients in buffer containing 0.1% NP-40. Gradients were centrifuged 22 h at 4°C in an SW41 rotor at 39,000 rpm and fractionated, and radioactivity was quantitated by scintillation counting. Fraction 1 represents the bottom of the gradient. The positions of the monomer (m), dimer (d), and E1 homotrimer (h) peaks are indicated. Recoveries ranged from 53 to 29%.
FIG. 5
Sucrose gradient sedimentation profiles of fus-1 and fus-1/ic viral spike proteins. 35S-labeled fus-1 or fus-1/ic virus was mixed with 1 mM liposomes and treated at the indicated pH for 10 min at 37°C. Samples were solubilized, neutralized, and analyzed by sucrose gradient sedimentation as for Fig. 4. Fraction 1 represents the bottom of the gradient. The positions of the monomer (m), dimer (d), and E1 homotrimer (h) peaks are indicated. Recoveries ranged from 32 to 63%.
FIG. 6
pH dependence of E1* conformational changes. (A) Reactivity with MAb E1a-1. 35S-labeled wt and fus-1 ectodomains were treated at the indicated pH for 10 min at 37°C in the presence of 1 mM liposomes, adjusted to neutral pH, solubilized in lysis buffer containing 1% Triton X-100, and immunoprecipitated with MAb E1a-1. E1* precipitation was quantitated by SDS-PAGE and phosphorimaging and compared to total E1* precipitated by a rabbit polyclonal antibody. Data represent the mean of three separate experiments. (B) E1* homotrimer formation. 35S-labeled wt SFV and fus-1 ectodomains were treated at the indicated pH for 10 min at 37°C in the presence of liposomes, adjusted to neutral pH, and solubilized in SDS sample buffer for 3 min at 30°C. E1 homotrimer formation was quantitated by SDS-PAGE and phosphorimaging. Data represent means of three separate experiments.
FIG. 7
Amino acid sequence differences among wt, mutant, and revertant virus structural proteins. The E2 isoleucine 12 change responsible for the fus-1 phenotype is marked (∗). No amino acid sequence differences were observed between wt/ic and wt SFV or between fus-1 and fus-1/ic. The positions of the SFV structural protein coding regions are marked.
FIG. 8
Sequence comparison of the E2 T12 region in alphaviruses. Amino acid numbering is given for SFV E2, starting with residue 1 at the amino terminus. The tetrabasic cleavage site which generates E3 and E2 is boxed, invariant residues in E2 are shown in bold, and sequence gaps are shown as dashes. The sequence of this region of fus-1 is identical to that of wt SFV except for the T12→I change. RRV, Ross River virus; CHICK (Chickungunya virus) (NCBI accession no. 576465); EEV, eastern equine encephalitis virus; OMN, O’Nyong-nyong virus; VEE, Venezuelan equine encephalitis virus; WEE, western equine encephalitis virus; SV, Sindbis virus; AURA, Aura virus; OCK, Ockelbo virus, as listed in references and .
Similar articles
- Role of conserved histidine residues in the low-pH dependence of the Semliki Forest virus fusion protein.
Qin ZL, Zheng Y, Kielian M. Qin ZL, et al. J Virol. 2009 May;83(9):4670-7. doi: 10.1128/JVI.02646-08. Epub 2009 Feb 25. J Virol. 2009. PMID: 19244325 Free PMC article. - Role of spike protein conformational changes in fusion of Semliki Forest virus.
Justman J, Klimjack MR, Kielian M. Justman J, et al. J Virol. 1993 Dec;67(12):7597-607. doi: 10.1128/JVI.67.12.7597-7607.1993. J Virol. 1993. PMID: 8230478 Free PMC article. - Biochemical consequences of a mutation that controls the cholesterol dependence of Semliki Forest virus fusion.
Chatterjee PK, Vashishtha M, Kielian M. Chatterjee PK, et al. J Virol. 2000 Feb;74(4):1623-31. doi: 10.1128/jvi.74.4.1623-1631.2000. J Virol. 2000. PMID: 10644331 Free PMC article. - Mechanisms of enveloped virus entry into cells.
Kielian M, Jungerwirth S. Kielian M, et al. Mol Biol Med. 1990 Feb;7(1):17-31. Mol Biol Med. 1990. PMID: 2182968 Review. - Assembly and entry mechanisms of Semliki Forest virus.
Garoff H, Wilschut J, Liljeström P, Wahlberg JM, Bron R, Suomalainen M, Smyth J, Salminen A, Barth BU, Zhao H, et al. Garoff H, et al. Arch Virol Suppl. 1994;9:329-38. doi: 10.1007/978-3-7091-9326-6_33. Arch Virol Suppl. 1994. PMID: 8032265 Review.
Cited by
- A tyrosine-to-histidine switch at position 18 of the Ross River virus E2 glycoprotein is a determinant of virus fitness in disparate hosts.
Jupille HJ, Medina-Rivera M, Hawman DW, Oko L, Morrison TE. Jupille HJ, et al. J Virol. 2013 May;87(10):5970-84. doi: 10.1128/JVI.03326-12. Epub 2013 Mar 20. J Virol. 2013. PMID: 23514884 Free PMC article. - Furin processing and proteolytic activation of Semliki Forest virus.
Zhang X, Fugère M, Day R, Kielian M. Zhang X, et al. J Virol. 2003 Mar;77(5):2981-9. doi: 10.1128/jvi.77.5.2981-2989.2003. J Virol. 2003. PMID: 12584323 Free PMC article. - Role of conserved histidine residues in the low-pH dependence of the Semliki Forest virus fusion protein.
Qin ZL, Zheng Y, Kielian M. Qin ZL, et al. J Virol. 2009 May;83(9):4670-7. doi: 10.1128/JVI.02646-08. Epub 2009 Feb 25. J Virol. 2009. PMID: 19244325 Free PMC article. - In vivo generation and characterization of a soluble form of the Semliki forest virus fusion protein.
Lu YE, Eng CH, Shome SG, Kielian M. Lu YE, et al. J Virol. 2001 Sep;75(17):8329-39. doi: 10.1128/jvi.75.17.8329-8339.2001. J Virol. 2001. PMID: 11483778 Free PMC article. - An Alphavirus E2 Membrane-Proximal Domain Promotes Envelope Protein Lateral Interactions and Virus Budding.
Byrd EA, Kielian M. Byrd EA, et al. mBio. 2017 Nov 7;8(6):e01564-17. doi: 10.1128/mBio.01564-17. mBio. 2017. PMID: 29114027 Free PMC article.
References
- Bentz J. Viral fusion mechanisms. Boca Raton, Fla: CRC Press; 1993. pp. 1–529.
- Binley J, Moore J P. HIV-cell fusion: the viral mousetrap. Nature. 1997;387:346–348. - PubMed
- Boggs W M, Hahn C S, Strauss E G, Strauss J H, Griffin D E. Low pH-dependent Sindbis virus-induced fusion of BHK cells: differences between strains correlate with amino acid changes in the E1 glycoprotein. Virology. 1989;169:485–488. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM52929/GM/NIGMS NIH HHS/United States
- T32 CA009173/CA/NCI NIH HHS/United States
- R01 GM052929/GM/NIGMS NIH HHS/United States
- 2T32 CA09173-15/CA/NCI NIH HHS/United States
- P30 CA013330/CA/NCI NIH HHS/United States
- P30-CA13330/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources