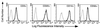HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A - PubMed (original) (raw)
HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A
N Lee et al. Proc Natl Acad Sci U S A. 1998.
Abstract
We previously showed that the availability of a nonamer peptide derived from certain HLA class I signal sequences is a necessary requirement for the stabilization of endogenous HLA-E expression on the surface of 721.221 cells. This led us to examine the ability of HLA-E to protect HLA class I transfectants from natural killer (NK) cell-mediated lysis. It was possible to implicate the CD94/NKG2A complex as an inhibitory receptor recognizing this class Ib molecule by using as target a .221 transfectant selectively expressing surface HLA-E. HLA-E had no apparent inhibitory effect mediated through the identified Ig superfamily (Ig-SF) human killer cell inhibitory receptors or ILT2/LIR1. Further studies of CD94/NKG2+ NK cell-mediated recognition of .221 cells transfected with different HLA class I allotypes (i.e., -Cw4, -Cw3, -B7) confirmed that the inhibitory interaction was mediated by CD94/NKG2A recognizing the surface HLA-E molecule, because only antibodies directed against either HLA-E, CD94, or CD94/NKG2A specifically restored lysis. Surface stabilization of HLA-E in cold-treated .221 cells loaded with appropriate peptides was sufficient to confer protection, resulting from recognition of the HLA class Ib molecule by the CD94/NKG2A inhibitory receptor. Consistent with the prediction that the ligand for CD94/NKG2A is expressed ubiquitously, our examination of HLA-E antigen distribution indicated that it is detectable on the surface of a wide variety of cell types.
Figures
Figure 1
Distribution of HLA-E in peripheral blood mononuclear cells. Peripheral blood lymphocytes were stained with phycoerythrin-labeled anti-CD3 (UCHT1, T cells), anti-CD20 (2H7, B cells), anti-CD14 (M5E2, monocytes), and anti-CD56 (B159, NK cells) mAbs and FITC-labeled anti-HLA-E (3D12) and FITC-labeled isotype-matched negative control (16G1). Cells were stained and analyzed as described in Materials and Methods, and relative fluorescent intensity was measured. Histograms of the FITC dimension of the four subpopulations are displayed separately as indicated in the upper right corner of each. Bold traces correspond to 3D12 and light traces correspond to the isotype-matched negative control.
Figure 2
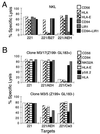
The presence of HLA-E on the surface of .221 cells inhibits NK lysis involving the CD94/NKG2 receptor. (A) Chromium release assay using line NKL (CD94/NKG2+, IgSF KIR− ILT2/LIR1+) against 721.221 and .221-AEH. Bars indicate the extent of lysis and are coded according to the presence of antibodies included in the respective assay (see key). (B) Chromium release assay using Z199+ (CD94/NKG2A+) NK clones against 721.221 and .221-AEH. Bars indicating extent of lysis are coded according to the antibody added (see key).
Figure 3
HLA-E does not prevent lysis through Ig-SF NK inhibitory receptors. (A) Line NKL was used in a chromium release assay against 721.221, .221-B27, and .221-AEH cells; the effects of anti-NK receptor mAbs, including the anti-ILT2/LIR1 antibody HP-F1, were tested. (B) Clones M3/17 and M3/5 were selected for their expression pattern of CD94/NKG2A and p58.2 (phenotype indicated in parentheses), determined by FACS analysis. Z199− cells not expressing CD94/NKG2A were not inhibited by HLA-E despite the presence of KIR-type receptor p58.2. In contrast, .221/AEH and .221-Cw3 cells inhibited the CD94/NKG2A+ p58.2- NK clones (Lower). In both cases, anti-HLA-E mAb reconstituted lysis comparably to anti-CD94/NKG2A. In both A and B, target cells are indicated beneath each and bars (keyed on right) indicate the extent of lysis.
Figure 4
Appropriate peptide binding to HLA-E is necessary for inhibition of NK lysis mediated through CD94/NKG2A. (A Left) Synthesized peptides were added to cold-treated .221-E cells, and the respective synthetic peptide was added as described in Materials and Methods. Histograms of FACS-analyzed cold-treated .221-E and .221-E plus peptide are presented. Bold traces correspond to the cells treated with peptide and semi-bold correspond to cells with no peptide added. The dotted trace in the experiment where no peptide was added (Top Left) corresponds to .221-E cells that were not cold-treated and thus lacking detectable HLA-E surface expression. The sequence of the peptide added and the name of a representative HLA allotype from which that peptide could be derived are indicated in the upper portion of each histogram. (Right) Chromium release assays using line NKL against the untreated and peptide-treated cells. The results are placed in the position corresponding to the respective FACS histogram on the left. Bars indicate the extent of lysis, and the respective antibody treatment is indicted beneath each bar. (B) Chromium release assays using NK clones (Z199+ EB6− ILT2/LIR1−) against .221 and .221 transfectants as targets. Clone names are indicated in the upper part of each box, and target cell names are indicated beneath each set of bars. Bars indicating extent of lysis are keyed (right) according to the antibody added.
Comment in
- HLA class I specificity for natural killer cell receptor CD94/NKG2A: two for one in more ways than one.
Yokoyama WM. Yokoyama WM. Proc Natl Acad Sci U S A. 1998 Apr 28;95(9):4791-4. doi: 10.1073/pnas.95.9.4791. Proc Natl Acad Sci U S A. 1998. PMID: 9560179 Free PMC article. No abstract available.
Similar articles
- HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C.
Braud VM, Allan DS, O'Callaghan CA, Söderström K, D'Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, Lanier LL, McMichael AJ. Braud VM, et al. Nature. 1998 Feb 19;391(6669):795-9. doi: 10.1038/35869. Nature. 1998. PMID: 9486650 - The ILT2(LIR1) and CD94/NKG2A NK cell receptors respectively recognize HLA-G1 and HLA-E molecules co-expressed on target cells.
Navarro F, Llano M, Bellón T, Colonna M, Geraghty DE, López-Botet M. Navarro F, et al. Eur J Immunol. 1999 Jan;29(1):277-83. doi: 10.1002/(SICI)1521-4141(199901)29:01<277::AID-IMMU277>3.0.CO;2-4. Eur J Immunol. 1999. PMID: 9933109 - HLA-G recognition by human natural killer cells. Involvement of CD94 both as inhibitory and as activating receptor complex.
Pende D, Sivori S, Accame L, Pareti L, Falco M, Geraghty D, Le Bouteiller P, Moretta L, Moretta A. Pende D, et al. Eur J Immunol. 1997 Aug;27(8):1875-80. doi: 10.1002/eji.1830270809. Eur J Immunol. 1997. PMID: 9295021 - Natural killer cell surveillance of intracellular antigen processing pathways mediated by recognition of HLA-E and Qa-1b by CD94/NKG2 receptors.
O'Callaghan CA. O'Callaghan CA. Microbes Infect. 2000 Apr;2(4):371-80. doi: 10.1016/s1286-4579(00)00330-0. Microbes Infect. 2000. PMID: 10817639 Review. - The CD94/NKG2 C-type lectin receptor complex: involvement in NK cell-mediated recognition of HLA class I molecules.
López-Botet M, Carretero M, Pérez-Villar J, Bellón T, Llano M, Navarro F. López-Botet M, et al. Immunol Res. 1997;16(2):175-85. doi: 10.1007/BF02786361. Immunol Res. 1997. PMID: 9212363 Review.
Cited by
- Single cell transcriptional zonation of human psoriasis skin identifies an alternative immunoregulatory axis conducted by skin resident cells.
Gao Y, Yao X, Zhai Y, Li L, Li H, Sun X, Yu P, Xue T, Li Y, Hu Y. Gao Y, et al. Cell Death Dis. 2021 May 6;12(5):450. doi: 10.1038/s41419-021-03724-6. Cell Death Dis. 2021. PMID: 33958582 Free PMC article. - T-cell tolerance in cancer.
Nurieva R, Wang J, Sahoo A. Nurieva R, et al. Immunotherapy. 2013 May;5(5):513-531. doi: 10.2217/imt.13.33. Immunotherapy. 2013. PMID: 23638746 Free PMC article. Review. - Herpes B virus, macacine herpesvirus 1, breaks simplex virus tradition via major histocompatibility complex class I expression in cells from human and macaque hosts.
Vasireddi M, Hilliard J. Vasireddi M, et al. J Virol. 2012 Dec;86(23):12503-11. doi: 10.1128/JVI.01350-12. Epub 2012 Sep 12. J Virol. 2012. PMID: 22973043 Free PMC article. - The Genetic Mechanisms Driving Diversification of the KIR Gene Cluster in Primates.
Bruijnesteijn J, de Groot NG, Bontrop RE. Bruijnesteijn J, et al. Front Immunol. 2020 Sep 11;11:582804. doi: 10.3389/fimmu.2020.582804. eCollection 2020. Front Immunol. 2020. PMID: 33013938 Free PMC article. Review. - Hypoimmunogenic human iPSCs expressing HLA-G, PD-L1, and PD-L2 evade innate and adaptive immunity.
Tsuneyoshi N, Hosoya T, Takeno Y, Saitoh K, Murai H, Amimoto N, Tatsumi R, Watanabe S, Hasegawa Y, Kikkawa E, Goto K, Nishigaki F, Tamura K, Kimura H. Tsuneyoshi N, et al. Stem Cell Res Ther. 2024 Jul 2;15(1):193. doi: 10.1186/s13287-024-03810-4. Stem Cell Res Ther. 2024. PMID: 38956724 Free PMC article.
References
- Colonna M, Samaridis J. Science. 1995;268:405–408. - PubMed
- Wagtmann N, Rajagopalan S, Winter C C, Peruzzi M, Long E O. Immunity. 1995;3:801–809. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous