Nuclear localization of IkappaB alpha is mediated by the second ankyrin repeat: the IkappaB alpha ankyrin repeats define a novel class of cis-acting nuclear import sequences - PubMed (original) (raw)
Nuclear localization of IkappaB alpha is mediated by the second ankyrin repeat: the IkappaB alpha ankyrin repeats define a novel class of cis-acting nuclear import sequences
S Sachdev et al. Mol Cell Biol. 1998 May.
Abstract
The ability of the IkappaB alpha protein to sequester dimeric NF-kappaB/Rel proteins in the cytoplasm provides an effective mechanism for regulating the potent transcriptional activation properties of NF-kappaB/Rel family members. IkappaB alpha can also act in the nucleus as a postinduction repressor of NF-kappaB/Rel proteins. The mechanism by which IkappaB alpha enters the nucleus is not known, as IkappaB alpha lacks a discernible classical nuclear localization sequence (NLS). We now report that nuclear localization of IkappaB alpha is mediated by a novel nuclear import sequence within the second ankyrin repeat. Deletion of the second ankyrin repeat or alanine substitution of hydrophobic residues within the second ankyrin repeat disrupts nuclear localization of IkappaB alpha. Furthermore, a region encompassing the second ankyrin repeat of IkappaB alpha is able to function as a discrete nuclear import sequence. The presence of a discrete nuclear import sequence in IkappaB alpha suggests that cytoplasmic sequestration of the NF-kappaB/Rel-IkappaB alpha complex is a consequence of the mutual masking of the NLS within NF-kappaB/Rel proteins and the import sequence within IkappaB alpha. Nuclear import may be a conserved property of ankyrin repeat domains (ARDs), as the ARDs from two other ARD-containing proteins, 53BP2 and GABPbeta, are also able to function as nuclear import sequences. We propose that the IkappaB alpha ankyrin repeats define a novel class of cis-acting nuclear import sequences.
Figures
FIG. 1
Domain organization of IκBα. The avian (p40) and mammalian (MAD3) IκBα proteins are represented by long rectangular boxes. The numbers to the left of each box indicate the first amino acid of each protein, and the numbers to the right of each box indicate the total number of amino acids in each protein. The IκBα proteins contain an N-terminal regulatory domain, a central domain containing five ankyrin repeats, and a C-terminal acidic and serine-rich (PEST) domain. The sites of N-terminal cytokine-inducible serine phosphorylation and the sites of constitutive serine phosphorylation within the C-terminal PEST domain of IκBα are indicated by the circled P’s. The amino acid sequences of two clusters of hydrophobic residues are indicated in the single-letter code below the rectangle representing each IκBα protein. The residues relevant to the present work are in boldface type, and the mutations introduced into the IκBα proteins are indicated. The scale of this line drawing is indicated by the length of the bar at the bottom of the figure.
FIG. 2
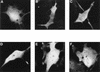
Cellular localization of wild-type and mutant IκBα proteins. CEF were transfected with SNV-derived retroviral vectors (A to D) and COS-1 cells were transfected with CMV-derived expression vectors (E to H) encoding either wild-type p40 (A and E), p40-114A3 (B and F), MAD3 (C and G), or MAD3-110A3 (D and H), or COS-1 cells were transfected with SNV-derived expression vectors encoding either p40-ΔAR2 (I), p40-ΔAR2+3 (J), p40-ΔAR4 (K), or p40-ΔAR5 (L). The p40-114A3 protein contains alanine substitutions for leucine 119, leucine 121, and isoleucine 124 in p40. The MAD3-110A3 protein contains alanine substitutions for leucine 115, leucine 117, and isoleucine 120 in MAD3. The p40-ΔAR2 protein contains a deletion of amino acids 98 to 142, encompassing the second ankyrin repeat in p40. The p40-ΔAR2+3 protein contains a deletion of amino acids 117 to 188, encompassing the second and third ankyrin repeats in p40. The p40-ΔAR4 protein contains a deletion of amino acids 189 to 222, encompassing the fourth ankyrin repeat in p40. The p40-ΔAR5 protein contains a deletion of amino acids 223 to 256, encompassing the fifth ankyrin repeat in p40. The cellular localization of the proteins in transfected cells was determined by indirect immunofluorescence with anti-p40 or anti-MAD3 serum. The cells shown are representative of more than 200 cells that were positive for the expression of the indicated proteins (see Table 1 for quantitation).
FIG. 3
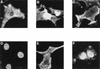
Cellular distribution of IκBα is independent of the p50, p52, p65 (RelA), and c-Rel proteins. WT+/+ (A), p50−/− (B), p65−/− (C), p50/p52−/− (D), and p50/p65−/− (E) mouse 3T3 fibroblasts or c-Rel/p65−/− primary MEF (F) were transfected with CMV-derived expression vectors coding for wild-type MAD3. The cellular localization of the ectopically expressed MAD3 protein was determined by indirect immunofluorescence with anti-MAD3 serum. The results shown are representative of at least 100 cells that were positive for expression of the wild-type MAD3 protein (see Table 2 for quantitation).
FIG. 4
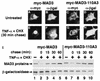
Nuclear localization of newly synthesized IκBα requires the integrity of the second ankyrin repeat. (A to H) COS-1 cells were cotransfected with CMV-derived expression vectors encoding β-galactosidase and either epitope-tagged MAD3 (myc-MAD3) or epitope-tagged MAD3-110A3 (myc-MAD3-110A3). At 36 h posttransfection, the transfected cells were either refed with complete medium (A to D), or were cultured in complete medium containing TNF-α (10 ng/ml) and CHX (100 μg/ml) for 4 h. The TNF-α and CHX were subsequently removed, and the transfected cells were chased in complete medium for 0, 15, 30, or 60 min. The localization of the β-galactosidase protein (B, D, F, and H) and either the myc-MAD3 (A and E) or the myc-MAD3-110A3 (C and G) protein was determined by anti-β-galactosidase (α-βgal) and anti-myc (α-myc) double-label indirect immunofluorescence, as indicated. The cellular localization of the ectopically expressed proteins at 30 min posttreatment is shown (E to H). The cells shown are representative of more than 25 cells that were positive for expression of both β-galactosidase and the respective myc-MAD3 protein (see Table 3 for quantitation). (I) For determination of the protein levels of the ectopically expressed proteins, cell lysates of the untreated and the TNF-α- and CHX-treated samples were collected in parallel. Equivalent amounts of each cell lysate were subjected to immunoblot analysis, and the levels of the ectopically expressed proteins from untreated samples (lanes 1 and 6) or from samples treated with TNF-α and CHX for 4 h and subsequently chased in complete medium for 0 (lanes 2 and 7), 15 (lanes 3 and 8), 30 (lanes 4 and 9), or 60 (lanes 5 and 10) min were determined by anti-MAD3 and anti-β-galactosidase immunoblots, as indicated. The myc-MAD3 and β-galactosidase proteins are indicated by arrows. Only the relevant portions of each immunoblot are shown.
FIG. 5
Nuclear import function of the second ankyrin repeat from IκB proteins. Fusion proteins between NPc and either the avian IκBα protein (p40), the mammalian IκBβ protein, the mammalian Bcl-3 protein, or the M9 nuclear import signal from hnRNP A1 are indicated (colons show fusions). The NPc protein comprises amino acids 2 through 150 of nucleoplasmin and contains an N-terminal epitope tag derived from the c-Myc protein. The ARD from p40, the second ankyrin repeat from p40 (p40-AR2), the second ankyrin repeat from IκBβ (IκBβ-AR2), the second ankyrin repeat from Bcl-3 (Bcl-3-AR2), or the M9 nuclear import signal was fused onto the C terminus of NPc. The following NPc-p40 fusion proteins were constructed with the indicated p40-derived amino acids (in parentheses): p40-ARD (49 to 272), p40-AR2 (103 to 149), p40-AR2-βN/αN/αC (103 to 138), p40-AR2-αN/αC/βC (117 to 149), and p40-AR2-αN (113 to 130). The IκBβ-derived amino acids used to construct the NPc–IκBβ-AR2 fusion protein were 83 to 128. The Bcl-3-derived amino acids used to construct the NPc–Bcl-3-AR2 fusion protein were 160 to 206. The hnRNP A1-derived amino acids used to construct the NPc-M9 fusion protein were 268 to 305. The cellular localization of each fusion protein was determined in COS-1 cells with anti-myc IgG. Cells that were positive for expression of the indicated fusion proteins were classified as having predominantly nuclear staining (N), staining that was distributed equally between the nucleus and the cytoplasm (N/C), or staining that was predominantly cytoplasmic (C). At least 200 cells that were positive for expression of each fusion protein were scored, and the percentage of cells in each category is given.
FIG. 6
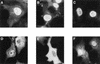
Nuclear localization of NPc upon fusion of the second ankyrin repeat of IκBα. COS-1 cells were transfected with CMV-based expression vectors encoding NPc (A), NPc–p40-ARD (B), NPc–p40-ARD-114A3 (C), NPc–p40-AR2 (D), NPc–p40-AR2-114A3 (E), or NPc-M9 (F). The NPc fusion proteins contain an N-terminal epitope tag derived from the c-Myc protein. The cellular localization of the NPc proteins in transfected cells was determined by indirect immunofluorescence with anti-myc IgG. The cells shown are representative of more than 200 cells that were positive for the expression of the indicated proteins.
FIG. 7
Nuclear import function of ankyrin repeat domains. (A) The structures of fusion proteins between NPc and the ARDs from the mammalian IκBα protein (MAD3), 53BP2, GABPβ, and Notch1 are indicated. The NPc protein comprises amino acids 2 through 150 of nucleoplasmin and contains an N-terminal epitope tag derived from the c-Myc protein. The ARDs from the indicated proteins were fused onto the C terminus of NPc. The amino acid residues from each protein that were used to construct each of the NPc-ARD fusion proteins are indicated. The cellular localization of the NPc-ARD fusion proteins was determined in COS-1 cells using anti-myc IgG. (B) The structure of a fusion protein between PK and the ARD from 53BP2 is indicated. Amino acids 17 through 524 of PK were used to construct the fusion protein, which also contains an N-terminal epitope tag derived from the c-Myc protein. The amino acid residues that were used to construct the PK–53BP2-ARD fusion protein are indicated. The cellular localization of the PK–53BP2-ARD fusion protein was determined in COS-1 cells using anti-myc IgG. For both panels, cells that were positive for expression of the indicated fusion proteins were classified as having predominantly nuclear staining (N), staining that was distributed equally between the nucleus and the cytoplasm (N/C), or staining that was predominantly cytoplasmic (C). At least 200 cells that were positive for expression of each fusion protein were scored, and the percent of cells in each category is given. Colons show fusions.
FIG. 8
Nuclear localization of NPc or PK upon fusion of ARDs. COS-1 cells were transfected with CMV-based expression vectors encoding NPc–MAD3-ARD (A), NPc–53BP2-ARD (B), NPc–GABPβ-ARD (C), NPc–Notch1-ARD (D), PK (E), or PK–53BP2-ARD (F). The NPc and PK fusion proteins contain an N-terminal epitope tag derived from the c-Myc protein. The cellular localization of the NPc and PK fusion proteins in transfected cells was determined by indirect immunofluorescence with anti-myc IgG. The cells shown are representative of more than 200 cells that were positive for the expression of the indicated proteins.
FIG. 9
Coimmunoprecipitation analysis of wild-type and mutant IκBα proteins. COS-1 cells were mock transfected (lane 4), transfected with a CMV-derived expression vector encoding p65 (RelA) (lanes 1 to 3), or transfected with a CMV-derived expression vector encoding c-Rel (lanes 5 to 7). CMV-derived expression vectors encoding either the wild-type LBD-p40 (lanes 2 and 6) or the LBD-p40-114A3 protein (lanes 3 and 7) were included in some transfections. Cell lysates were subjected to immunoprecipitation with affinity-purified anti-LBD rabbit IgG followed by immunoblot analysis with either anti-p65 mouse IgG (top panels, lanes 1 to 3), or anti-c-Rel mouse IgG (top panels, lanes 4 to 7). Cell lysates were also subjected to direct immunoblot analysis with anti-p65 rabbit serum (middle panels, lanes 1 to 3), with anti-c-Rel rabbit serum (middle panels, lanes 4 to 7), or with anti-p40 rabbit serum (bottom panels, lanes 1 to 7). The p65 (RelA), c-Rel, and p40 proteins are indicated by arrows. Only the relevant portions of each immunoblot are shown.
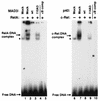
FIG. 10
Inhibitory properties of wild-type and mutant IκBα proteins. COS-1 cells were mock transfected (lanes 1 and 6) or were transfected with CMV-derived expression vectors encoding either p65 (RelA) (lanes 2 to 5) or c-Rel (lanes 7 to 10). CMV-derived expression vectors encoding wild-type MAD3 (lane 3), mutant MAD3-110A3 (lane 4), wild-type p40 (lane 8), or mutant p40-114A3 (lane 9) were included in some transfections. Cell lysates were analyzed for proteins that bound to a 32P-labeled oligonucleotide containing a palindromic κB site. A 100-fold excess of the unlabeled palindromic κB oligonucleotide was included in some DNA-binding reaction mixtures (lanes 5 and 10). The DNA-binding reaction mixtures were electrophoresed through a 5% nondenaturing polyacrylamide gel. The positions of the respective Rel-DNA complexes and unbound DNA are indicated by arrows. The endogenous Rel DNA-binding activity in COS-1 cells is denoted by an asterisk.
References
- Arenzana-Seisdedos F, Turpin P, Rodriguez M, Thomas D, Hay R T, Virelizier J-L, Dargemont C. Nuclear localization of IκBα promotes active transport of NF-κB from the nucleus to the cytoplasm. J Cell Sci. 1997;110:369–378. - PubMed
- Baldwin A S., Jr The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–681. - PubMed
- Beg A A, Ruben S M, Scheinman R I, Haskill S, Rosen C A, Baldwin A S., Jr IκB interacts with the nuclear localization sequences of the subunits of NF-κB: a mechanism for cytoplasmic retention. Genes Dev. 1992;6:1899–1913. - PubMed
- Beg A A, Baldwin A S., Jr The IκB proteins: multifunctional regulators of Rel/NF-κB in the immune system. Genes Dev. 1993;7:2064–2070. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases