Transcriptional activation of the integrated chromatin-associated human immunodeficiency virus type 1 promoter - PubMed (original) (raw)
Transcriptional activation of the integrated chromatin-associated human immunodeficiency virus type 1 promoter
A El Kharroubi et al. Mol Cell Biol. 1998 May.
Abstract
The regulation of human immunodeficiency virus type 1 (HIV-1) gene expression involves a complex interplay between cellular transcription factors, chromatin-associated proviral DNA, and the virus-encoded transactivator protein, Tat. Here we show that Tat transactivates the integrated HIV-1 long terminal repeat (LTR), even in the absence of detectable basal promoter activity, and this transcriptional activation is accompanied by chromatin remodeling downstream of the transcription initiation site, as monitored by increased accessibility to restriction endonucleases. However, with an integrated promoter lacking both Sp1 and NF-kappaB sites, Tat was unable to either activate transcription or induce changes in chromatin structure even when it was tethered to the HIV-1 core promoter upstream of the TATA box. Tat responsiveness was observed only when Sp1 or NF-kappaB was bound to the promoter, implying that Tat functions subsequent to the formation of a specific transcription initiation complex. Unlike Tat, NF-kappaB failed to stimulate the integrated transcriptionally silent HIV-1 promoter. Histone acetylation renders the inactive HIV-1 LTR responsive to NF-kappaB, indicating that a suppressive chromatin structure must be remodeled prior to transcriptional activation by NF-kappaB. Taken together, these results suggest that Sp1 and NF-kappaB are required for the assembly of transcriptional complexes on the integrated viral promoter exhibiting a continuum of basal activities, all of which are fully responsive to Tat.
Figures
FIG. 1
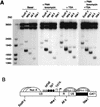
Effects of NF-κB and Sp1 on the activity of integrated HIV-1 LTRs. (A) Schematic representation of the WT-, KBmt-, and DS-LTR CAT constructs. The LTR (U3, R, and U5) and gag leader sequence (GLS) are represented by open rectangles. Binding sites for the NF-κB, Sp1, TFIID (TATA box), AP-1, AP-3-like, and DBF1 transcription factors are shown. The transcription initiation site is indicated by the arrow at the junction between in U3 and R sites. In the DS-LTR, the regulatory elements downstream of the transcription start site (AP-1, AP-3, DBF1, and the downstream Sp1) were deleted and an unrelated sequence (filled rectangle) was inserted between U5 and the CAT gene. (B) CAT activities in unstimulated (Basal) or transfected (1 μg of Sp1 or NF-κB expression vector) WT3, KB1, and DS2 cells. (C) HeLa cells were transiently transfected with WT-, DS- or KBmt-LTR-CAT reporter plasmid alone (Basal) or along with 1 μg of vector expressing Sp1 or NF-κB as indicated at the bottom. At 24 h posttransfection, CAT activity was determined in cell lysates and expressed as percent acetylated chloramphenicol (% acetylation). The bars show the results from a representative transfection experiment; the numbers above the bars indicate average fold induction.
FIG. 2
Synergistic activation of the integrated HIV-1 promoter by treatment of cells with histone deacetylase inhibitor TSA and NF-κB inducers. WT3, DS2, and KB1 cells were untreated (−) or treated (+) with PMA plus ionomycin, TNFα, and/or TSA as indicated at the bottom. At 36 h postinduction, CAT activity was determined in cell lysates and expressed as percent acetylated chloramphenicol (% acetylation). The bars show the results from a representative transfection experiment; the numbers above the bars indicate average fold induction.
FIG. 3
NF-κB stimulation induces increased accessibility of the integrated HIV-1 WT-LTR to restriction endonucleases. (A and B) Nuclei isolated from WT3 and KB1 cells maintained in DMEM (−) or stimulated with PMA plus ionomycin (+) were digested with the restriction enzymes indicated at the top. DNA was then purified, digested to completion with _Bgl_I and _Hin_cII, and analyzed by indirect end labeling. A molecular weight size marker was prepared by digesting a previously described LTR-CAT plasmid, pLCH (15), with several restriction enzymes. (C) Schematic representation of the putative nucleosomal structure and partial restriction map of WT-LTR. Nuc A and Nuc C are positioned relative to micrococcal nuclease and restriction enzyme cleavage sites (11). Nuc A is shown as three adjacent and overlapping structures to indicate that it has not been precisely positioned. Nuc C is represented by a hatched structure to indicate that its accessibility to endonucleases is increased following stimulation with PMA plus ionomycin. GLS, gag leader sequence.
FIG. 4
TSA treatment of DS2 cells enhances endonuclease accessibility of the integrated LTR. Nuclei isolated from DS2 cells maintained in DMEM (Basal) or treated with either PMA plus ionomycin, TSA, or a combination of both PMA, ionomycin, and TSA, as indicated, were digested with the restriction enzymes indicated at the top. DNA was then purified, digested to completion with _Bgl_I and _Hin_cII, and analyzed by indirect end labeling. (B) Schematic representation of the putative nucleosomal structure and partial restriction map of DS-LTR. Nuc C and Nuc D are represented by a hatched structure to indicate that their accessibility to endonucleases is increased following treatment with TSA.
FIG. 5
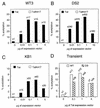
Tat strongly transactivates the integrated HIV-1 LTR. (A to C) Titration of CAT activities in WT3 (A), DS2 (B), and KB1 (C) cell clones transfected with either pSV-Tat (Tat) or pSV-TatK41T (TatK41T). (D) HeLa cells were transiently transfected with WT- or DS-LTR-CAT reporter plasmid and increasing amounts of Tat expression vector as indicated at the bottom. At 24 h posttransfection, CAT activity was determined in cell lysates and expressed as percent acetylated chloramphenicol (% acetylation). The bars show the results from a representative transfection experiment; the numbers above the bars indicate average fold induction.
FIG. 6
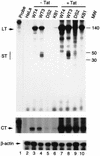
Tat stimulates transcription directed by the integrated HIV-1 LTR. RNase protection assays showing the levels of RNA synthesis in untransfected (− Tat) or transfected (+ Tat) HeLa cell clones carrying integrated copies of the WT-LTR (WT3 and WT4), the DS-LTR (DS2), or the KBmt-LTR (KB1). The probes used were (i) an HIV-1 LTR probe that detects both the short (ST) and long (LT) transcripts, (ii) a CAT probe to detect CAT transcripts that extend 680 nt downstream of the transcription start site (CT), and (iii) a β-actin probe that detects cellular actin mRNAs. Molecular weights (MW) are indicated in nucleotides.
FIG. 7
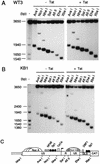
Accessibility of the integrated HIV-1 WT- and KBmt-LTRs to restriction endonucleases in the presence or absence of Tat. (A and B) Nuclei isolated from WT3 (A) and KB1 (B) cells maintained in DMEM (− Tat) or transfected with a Tat expression vector (+ Tat) were digested with the restriction enzymes indicated at the top. DNA was then purified, digested to completion with _Bgl_I and _Hin_cII and analyzed by indirect end labeling. A molecular weight size marker was prepared by digesting a previously described LTR-CAT plasmid, pLCH (15), with several restriction enzymes. (C) Schematic representation of the putative nucleosomal structure and partial restriction map of WT-LTR. Nuc C is represented by a hatched structure to indicate that its accessibility to endonucleases is increased following the expression of Tat protein. GLS, gag leader sequence.
FIG. 8
Chromatin changes associated with Tat expression in DS2 cells. (A) Nuclei isolated from cloned DS2 HeLa cells maintained in DMEM (− Tat) or transfected with either a Tat expression vector (+ Tat) or the Tat(K41T) expression vector (+ Tat K41T) were digested with the restriction enzymes indicated at the top. DNA was then purified, digested to completion with _Bgl_I and _Hin_cII, and analyzed by indirect end labeling. UD, undigested parental fragment; #, an unrelated band that hybridizes to the probe but lacks any HIV-1 LTR sequences. (B) Schematic representation of the putative nucleosomal structure of DS-LTR. Nuc C and Nuc D are represented as hatched structures to indicate that their structure is remodeled during Tat activation.
FIG. 9
Transcriptional activation and chromatin remodeling of the IRF-LTR. (A) Schematic representation of the putative nucleosomal structure of the IRF-LTR (14a). Nuc A, Nuc B, and Nuc C are represented as overlapping structures to indicate that their positions have not been precisely determined. The Nuc C segment is hatched to indicate that this region of chromatin is accessible to _Afl_II when the promoter is active. GLS, gag leader sequence. CAT activities in HeLa cells transiently transfected with the WT- or IRF-LTR-CAT reporter plasmid are shown at the right. (B) Left, HeLa cells carrying a single copy of the IRF-LTR were transfected with increasing amounts of Tat, Tat-IRF2, or IRF2-Tat expression vector, and CAT activities in cell lysates were determined. Right, IRF-LTR containing cells were transfected with increasing amounts of a vector expressing the IRF2-p65 fusion protein alone (−Tat) or along with 1 μg of Tat expressing vector (+Tat), and CAT activities were determined. (C) Cells carrying the IRF-LTR template were maintained in DMEM (Basal) or transfected with the indicated expression vectors. At 12 h posttransfection, nuclei were isolated and digested with _Ava_I, _Nhe_I, _Afl_II, and _Xba_I. DNA was purified and analyzed by indirect end labeling.
Similar articles
- Semen Exosomes Promote Transcriptional Silencing of HIV-1 by Disrupting NF-κB/Sp1/Tat Circuitry.
Welch JL, Kaddour H, Schlievert PM, Stapleton JT, Okeoma CM. Welch JL, et al. J Virol. 2018 Oct 12;92(21):e00731-18. doi: 10.1128/JVI.00731-18. Print 2018 Nov 1. J Virol. 2018. PMID: 30111566 Free PMC article. - Lack of responsiveness of a nuclear factor-kappaB-regulated promoter to transactivation by human immunodeficiency virus 1 Tat in HeLa cells.
Kelly GD, Morris CB, Offermann MK. Kelly GD, et al. Virology. 1999 Oct 10;263(1):128-38. doi: 10.1006/viro.1999.9966. Virology. 1999. PMID: 10544088 - The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation.
Jordan A, Defechereux P, Verdin E. Jordan A, et al. EMBO J. 2001 Apr 2;20(7):1726-38. doi: 10.1093/emboj/20.7.1726. EMBO J. 2001. PMID: 11285236 Free PMC article. - Chromatin-associated regulation of HIV-1 transcription: implications for the development of therapeutic strategies.
Quivy V, De Walque S, Van Lint C. Quivy V, et al. Subcell Biochem. 2007;41:371-96. Subcell Biochem. 2007. PMID: 17484137 Review. - Multiple modes of transcriptional regulation by the HIV-1 Tat transactivator.
Marcello A, Zoppé M, Giacca M. Marcello A, et al. IUBMB Life. 2001 Mar;51(3):175-81. doi: 10.1080/152165401753544241. IUBMB Life. 2001. PMID: 11547919 Review.
Cited by
- The histone chaperone protein Nucleosome Assembly Protein-1 (hNAP-1) binds HIV-1 Tat and promotes viral transcription.
Vardabasso C, Manganaro L, Lusic M, Marcello A, Giacca M. Vardabasso C, et al. Retrovirology. 2008 Jan 28;5:8. doi: 10.1186/1742-4690-5-8. Retrovirology. 2008. PMID: 18226242 Free PMC article. - Repression of Human T-lymphotropic virus type 1 Long Terminal Repeat sense transcription by Sp1 recruitment to novel Sp1 binding sites.
Fauquenoy S, Robette G, Kula A, Vanhulle C, Bouchat S, Delacourt N, Rodari A, Marban C, Schwartz C, Burny A, Rohr O, Van Driessche B, Van Lint C. Fauquenoy S, et al. Sci Rep. 2017 Mar 3;7:43221. doi: 10.1038/srep43221. Sci Rep. 2017. PMID: 28256531 Free PMC article. - HTLV-1 Tax mediated downregulation of miRNAs associated with chromatin remodeling factors in T cells with stably integrated viral promoter.
Rahman S, Quann K, Pandya D, Singh S, Khan ZK, Jain P. Rahman S, et al. PLoS One. 2012;7(4):e34490. doi: 10.1371/journal.pone.0034490. Epub 2012 Apr 4. PLoS One. 2012. PMID: 22496815 Free PMC article. - Epigenetic regulation of high glucose-induced proinflammatory cytokine production in monocytes by curcumin.
Yun JM, Jialal I, Devaraj S. Yun JM, et al. J Nutr Biochem. 2011 May;22(5):450-8. doi: 10.1016/j.jnutbio.2010.03.014. Epub 2010 Jul 22. J Nutr Biochem. 2011. PMID: 20655188 Free PMC article.
References
- Adams M, Sharmeen L, Kimpton J, Romeo J M, Garcia J V, Peterlin B M, Groudine M, Emerman M. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc Natl Acad Sci USA. 1994;91:3862–3866. - PMC - PubMed
- Archer T K, Lefebvre P, Wolford R G, Hager G L. Transcription factor loading on the MMTV promoter: a bidominal mechanism for promoter activation. Science. 1992;255:1573–1576. - PubMed
- Berkhout B, Gatignol A, Rabson A B, Jeang K-T. TAR-independent activation of the HIV-1 LTR: evidence that Tat requires specific regions of the promoter. Cell. 1990;62:757–767. - PubMed
- Berkhout B, Silverman R H, Jeang K-T. Tat transactivates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources