Yeast RNA polymerase II transcription in vitro is inhibited in the presence of nucleotide excision repair: complementation of inhibition by Holo-TFIIH and requirement for RAD26 - PubMed (original) (raw)
Yeast RNA polymerase II transcription in vitro is inhibited in the presence of nucleotide excision repair: complementation of inhibition by Holo-TFIIH and requirement for RAD26
Z You et al. Mol Cell Biol. 1998 May.
Abstract
The Saccharomyces cerevisiae transcription factor IIH (TFIIH) is essential both for transcription by RNA polymerase II (RNAP II) and for nucleotide excision repair (NER) of damaged DNA. We have established cell extracts which support RNAP II transcription from the yeast CYC1 promoter or NER of transcriptionally silent damaged DNA on independent plasmid templates and substrates. When plasmid templates and substrates for both processes are simultaneously incubated with these extracts, transcription is significantly inhibited. This inhibition is strictly dependent on active NER and can be complemented with purified holo-TFIIH. These results suggest that in the presence of active NER, TFIIH is preferentially mobilized from the basal transcription machinery for use in NER. Inhibition of transcription in the presence of active NER requires the RAD26 gene, the yeast homolog of the human Cockayne syndrome group B gene (CSB).
Figures
FIG. 1
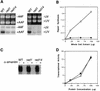
(A) NER in wild-type (strain W303-1B) and rad1 and rad14 deletion mutant extracts. NER was performed with 50 μg of extract at 25°C for 60 min. WT, wild type; +AAF, pUC18 DNA containing AAF adducts; −AAF, undamaged pGEM3Zf(+) DNA; +UV, UV-irradiated pUC18 DNA; −UV, undamaged pGEM3Zf(+) DNA. Ethidium bromide-stained gels (top) and autoradiograms of the gels (bottom) are shown. (B) Protein concentration dependence of DNA repair synthesis in cell extracts. AAF-damaged pUC18 DNA was incubated with the indicated amounts of yeast wild-type (strain W303-1B) or rad14 deletion mutant extracts for 1 h at 25°C. Quantitation was performed with a PhosphorImager. Open squares, wild type; closed diamonds, rad14 mutant. (C) RNAP II transcription of plasmid CYC1/G− was performed with 50 μg of extract at 25°C for 45 min. α-Amanitin (10 μg/ml) was included (+) or not included (−) as indicated over the gel. (D) Protein concentration dependence of transcription in cell extracts. Transcription of the template (200 ng) was performed at 25°C for 45 min with the indicated amounts of yeast wild-type (strain W303-1B) or rad14 mutant extracts. Quantitation was performed with a PhosphorImager. Open circles, wild type; closed squares, rad14 mutant.
FIG. 2
(A) Inhibition of RNAP II transcription in the presence of NER in yeast wild-type (strain W303-1B) extracts. After 20 min of preincubation at 20°C, transcription and NER were initiated by supplying appropriate reaction buffers (see Materials and Methods). Transcription of the CYC1/G− template (200 ng) was at 25°C for 45 min with 20 μg of cell extract in the presence (+) or absence (−) of NER substrates (pUC-AAF DNA and pUC-UV DNA). NER substrates (400 ng) and control pUC18 DNA (400 ng) were included as indicated over the gels. The 350- to 375-nucleotide transcripts were resolved on a 6% polyacrylamide–7 M urea gel and detected by phosphorimaging. (B) Transcription from the CYC1/G− template (200 ng) was carried out with 20 μg of various rad mutant extracts in the presence (+) or absence (−) of 400 ng of NER substrate or control pUC18 DNA as indicated over the gels. Incubations were at 25°C for 45 min. pUC-AAF, pUC18 DNA treated with AAAF; pUC-UV, UV-irradiated pUC18 DNA; pUC, pUC18 DNA.
FIG. 3
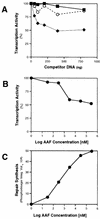
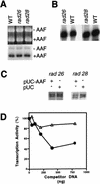
(A) Transcription inhibition in the presence of NER. Transcription of the CYC1/G− template (200 ng) was performed at 25°C for 45 min with 20 μg of extract in the presence of increasing amounts of pUC18 DNA (50, 100, 200, 400, or 800 ng) treated with 30 μM AAAF (pUC18-AAF), or with the same amount of undamaged pUC18 DNA. Quantitation was performed by phosphorimaging. Transcription in the absence of competitor DNA was normalized to 100%. Open circles, wild-type yeast strain with competitor pUC18 DNA; closed diamonds, wild-type strain with competitor pUC18-AAF DNA; closed squares, rad14 mutant with competitor pUC18-AAF DNA. (B) Transcription of the CYC1/G− template (200 ng) was performed with 20 μg of wild-type extract in the presence of pUC18-AAF DNA (400 ng) treated with increasing amounts of AAAF (30 nM to 3 mM) at 25°C for 45 min. Transcription in the presence of untreated pUC18 DNA was normalized to 100%. (C) NER was performed with 20 μg of wild-type extract in the presence of 400 ng of pUC18 DNA treated with increasing amounts of AAAF (30 nM to 3 mM), at 25°C for 60 min. Quantitation of repair synthesis was performed by phosphorimaging.
FIG. 4
Inhibition of RNAP II transcription from the CYC1/G− template requires active NER. (A) Transcription template (200 ng), pUC18 DNA treated with AAAF (pUC-AAF) DNA (400 ng), and 20 μg of wild-type extract were incubated at 20°C for 20 min. Reaction buffer (lacking CTP and UTP) was added, and NER was allowed to proceed at 25°C for the times indicated, following which CTP and UTP were added to initiate transcription for 45 min. pUC18 DNA (400 ng) was used instead of pUC-AAF DNA in the control experiment, and the amount of transcription was normalized to 1.0. (B) Transcription of CYC1/G− template (200 ng) was performed with 20 μg of wild-type extract in the presence (+) of pUC-AAF or pUC18 DNA (400 ng) at 25°C for 45 min. Deoxynucleoside triphosphates (dNTP’s) were included (+) or omitted (−) as indicated. (C) NER was performed with 20 μg of rad4, rad14, or a mixture of rad4 and rad14 mutant extracts under transcription-NER reaction conditions. +AAF, pUC18 DNA treated with AAAF; −AAF, undamaged pGEM3Zf(+) DNA. (D) Inhibition of transcription from the CYC1/G− template (200 ng) was restored to mixtures of rad4 and rad14 mutant extracts in the presence of the NER substrate (pUC18 DNA treated with UV irradiation). (E) Quantitation of the results shown in panel D. Quantitation was performed by phosphorimaging and normalized in the rad4 experiment to 1.0.
FIG. 5
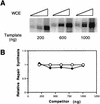
(A) Increased transcription from the CYC1/G− template by increasing the amount of template or extract. Transcription was performed at 25°C for 45 min with various amounts (10, 20, and 50 μg [indicated by the height of the large open triangle over each set of three lanes]) of wild-type extract in the presence of various amounts (200, 600, or 1,000 ng) of template. (B) NER was performed at 25°C for 60 min with 20 μg of wild-type extract in the presence of various amounts (200, 400, 600, 800, or 1,000 ng) of the transcription plasmid CYC1/G− (closed diamonds) or control undamaged pGEM3Zf(+) DNA (open circles). Repair synthesis was quantitated by phosphorimaging, and the level of repair synthesis in the presence of pGEM3Zf(+) DNA was normalized to 1.0. WCE, whole-cell extract.
FIG. 6
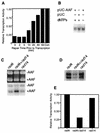
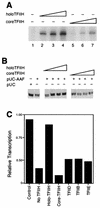
Inhibition of RNAP II transcription in the presence of NER is relieved by holo-TFIIH. (A) Both core TFIIH (lanes 2 to 4) and holo-TFIIH (lanes 5 to 7) restored transcription activity to a heat-inactivated nuclear extract (HNE). All reaction mixtures contained 6 μl of HNE and 70 ng of recombinant yeast TBP. Core and holo-TFIIH (0.5, 1.0, and 2.0 μl of Mono Q fractions [indicated by the height of the large open triangle over each set of three lanes]) were assayed. (B) Incubations contained CYC1/G− template (200 ng), pUC18 treated with AAAF (pUC-AAF) or control pUC18 DNA (400 ng), 20 μg of wild-type extract, and various amounts of holo- or core TFIIH (0.5, 1.0, 2.0, and 3.0 μl of MonoQ fraction [increasing amount of TFIIH indicated by the height of the large open triangle over each set of four lanes]). Incubations were at 20°C for 20 min, following which transcription-NER reaction buffer was added and incubation continued at 25°C for 45 min. (C) Reaction mixtures containing CYC1/G− template (200 ng), pUC-AAF or pUC plasmid (400 ng; control), and 20 μg of wild-type extract were supplemented with either distilled water, holo-TFIIH, core TFIIH, TFIIB (200 ng), TBP (200 ng), or TFIIE (200 ng). Incubations were at 20°C for 20 min, following which transcription-NER reaction buffer was added and incubation was continued at 25°C for 45 min. Quantitation of transcription was performed by phosphorimaging and normalized in the control experiment to 1.0.
FIG. 7
Inhibition of RNAP II transcription in the presence of NER requires RAD26 but not RAD28. (A) NER was performed with 50 μg of wild-type (WT), rad26, or rad28 extract in the presence of pUC18 treated with AAAF (pUC-AAF), pGEM3Zf(+) at 25°C for 60 min. −AAF, undamaged pGEM3Zf(+) DNA; +AAF, AAAF-treated pUC18 DNA. An ethidium bromide-stained gel (top) and an autoradiogram of the gel (bottom) are shown. (B) Transcription was with 50 μg of wild-type, rad26, or rad28 extract in the presence of CYC1/G− template at 25°C for 45 min. (C) Transcription from the CYC1/G− template was at 25°C for 45 min with 20 μg of rad26 or rad28 mutant extract in the presence (+) or absence (−) of pUC-AAF or control pUC plasmid (400 ng). (D) Transcription from the CYC1/G− template with 20 μg of rad26 or rad28 mutant extract was at 25°C for 45 min in the presence of various amounts of NER substrate (pUC-AAF). Transcription was quantitated as described above. Transcription in the absence of competitor DNA was normalized to 100%. Open triangles, rad26 extract; closed circles, rad28 extract.
FIG. 8
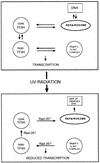
Model for RNAP II transcription inhibition in the presence of active NER. The top panel portrays an equilibrium steady state which determines the distribution of holo- and core TFIIH in RNAP II transcription initiation and NER complexes, respectively, in the absence of exogenous base damage to DNA. The lower panel shows that when cells are exposed to DNA-damaging agents, such as UV radiation, this steady state is shifted such that holo-TFIIH is mobilized out of RNAP II transcription initiation complexes and core TFIIH is appropriated for active NER. This process requires Rad26 protein.
Similar articles
- A yeast whole cell extract supports nucleotide excision repair and RNA polymerase II transcription in vitro.
Wang Z, Wu X, Friedberg EC. Wang Z, et al. Mutat Res. 1996 Sep 2;364(1):33-41. doi: 10.1016/0921-8777(96)00019-5. Mutat Res. 1996. PMID: 8814336 - The yeast TFB1 and SSL1 genes, which encode subunits of transcription factor IIH, are required for nucleotide excision repair and RNA polymerase II transcription.
Wang Z, Buratowski S, Svejstrup JQ, Feaver WJ, Wu X, Kornberg RD, Donahue TF, Friedberg EC. Wang Z, et al. Mol Cell Biol. 1995 Apr;15(4):2288-93. doi: 10.1128/MCB.15.4.2288. Mol Cell Biol. 1995. PMID: 7891722 Free PMC article. - Different forms of TFIIH for transcription and DNA repair: holo-TFIIH and a nucleotide excision repairosome.
Svejstrup JQ, Wang Z, Feaver WJ, Wu X, Bushnell DA, Donahue TF, Friedberg EC, Kornberg RD. Svejstrup JQ, et al. Cell. 1995 Jan 13;80(1):21-8. doi: 10.1016/0092-8674(95)90447-6. Cell. 1995. PMID: 7813015 - Ten years of TFIIH.
Coin F, Egly JM. Coin F, et al. Cold Spring Harb Symp Quant Biol. 1998;63:105-10. doi: 10.1101/sqb.1998.63.105. Cold Spring Harb Symp Quant Biol. 1998. PMID: 10384274 Review. No abstract available. - Transcription coupled nucleotide excision repair in the yeast Saccharomyces cerevisiae: The ambiguous role of Rad26.
Li S. Li S. DNA Repair (Amst). 2015 Dec;36:43-48. doi: 10.1016/j.dnarep.2015.09.006. Epub 2015 Sep 10. DNA Repair (Amst). 2015. PMID: 26429063 Review.
Cited by
- Local UV-induced DNA damage in cell nuclei results in local transcription inhibition.
Moné MJ, Volker M, Nikaido O, Mullenders LH, van Zeeland AA, Verschure PJ, Manders EM, van Driel R. Moné MJ, et al. EMBO Rep. 2001 Nov;2(11):1013-7. doi: 10.1093/embo-reports/kve224. EMBO Rep. 2001. PMID: 11713193 Free PMC article. - Preparation of efficient excision repair competent cell-free extracts from C. reinhardtii cells.
Chaudhari V, Raghavan V, Rao BJ. Chaudhari V, et al. PLoS One. 2014 Oct 9;9(10):e109160. doi: 10.1371/journal.pone.0109160. eCollection 2014. PLoS One. 2014. PMID: 25299516 Free PMC article. - Tfb5 is partially dispensable for Rad26 mediated transcription coupled nucleotide excision repair in yeast.
Ding B, Ruggiero C, Chen X, Li S. Ding B, et al. DNA Repair (Amst). 2007 Nov;6(11):1661-9. doi: 10.1016/j.dnarep.2007.06.001. Epub 2007 Jul 20. DNA Repair (Amst). 2007. PMID: 17644494 Free PMC article. - Rous-Whipple Award Lecture. Nucleotide excision repair and cancer predisposition: A journey from man to yeast to mice.
Friedberg EC. Friedberg EC. Am J Pathol. 2000 Sep;157(3):693-701. doi: 10.1016/s0002-9440(10)64581-6. Am J Pathol. 2000. PMID: 10980107 Free PMC article. No abstract available.
References
- Bardwell A J, Bardwell L, Tomkinson A E, Friedberg E C. Specific cleavage of model recombination and repair intermediates by the yeast RAD1-RAD10 DNA endonuclease. Science. 1994;265:2082–2085. - PubMed
- Bohr V A, Smith C A, Okumoto D S, Hanawalt P C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous