Use of inducible feedback-resistant N-acetylglutamate synthetase (argA) genes for enhanced arginine biosynthesis by genetically engineered Escherichia coli K-12 strains - PubMed (original) (raw)
Use of inducible feedback-resistant N-acetylglutamate synthetase (argA) genes for enhanced arginine biosynthesis by genetically engineered Escherichia coli K-12 strains
B S Rajagopal et al. Appl Environ Microbiol. 1998 May.
Abstract
The goal of this work was to construct Escherichia coli strains capable of enhanced arginine production. The arginine biosynthetic capacity of previously engineered E. coli strains with a derepressed arginine regulon was limited by the availability of endogenous ornithine (M. Tuchman, B. S. Rajagopal, M. T. McCann, and M. H. Malamy, Appl. Environ. Microbiol. 63:33-38, 1997). Ornithine biosynthesis is limited due to feedback inhibition by arginine of N-acetylglutamate synthetase (NAGS), the product of the argA gene and the first enzyme in the pathway of arginine biosynthesis in E. coli. To circumvent this inhibition, the argA genes from E. coli mutants with feedback-resistant (fbr) NAGS were cloned into plasmids that contain "arg boxes," which titrate the ArgR repressor protein, with or without the E. coli carAB genes encoding carbamyl phosphate synthetase and the argI gene for ornithine transcarbamylase. The free arginine production rates of "arg-derepressed" E. coli cells overexpressing plasmid-encoded carAB, argI, and fbr argA genes were 3- to 15-fold higher than that of an equivalent system overexpressing feedback-sensitive wild-type (wt) argA. The expression system with fbr argA produced 7- to 35-fold more arginine than a system overexpressing carAB and argI genes on a plasmid in a strain with a wt argA gene on the chromosome. The arginine biosynthetic capacity of arg-derepressed DH5 alpha strains with plasmids containing only the fbr argA gene was similar to that of cells with plasmids also containing the carAB and argI genes. Plasmids containing wt or fbr argA were stably maintained under normal growth conditions for at least 18 generations. DNA sequencing identified different point mutations in each of the fbr argA mutants, specifically H15Y, Y19C, S54N, R58H, G287S, and Q432R.
Figures
FIG. 1
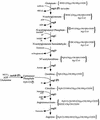
Pathway of arginine biosynthesis in E. coli. (P), overexpression of the genes on engineered plasmids; wt, feedback sensitive; fbr, feedback resistant; AcSCoA, acetyl coenzyme A; HSCoA, coenzyme A.
FIG. 2
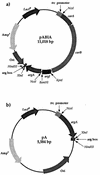
pABIA (linked as an operon) (a) and pA vectors (b) engineered for this study. The vectors contained the gene(s) downstream from a control region which includes the trc promoter and a lac operator. The constructs also contained an arg box cloned from the argI gene for binding and titration of the arginine repressor. Ori, ColE1 replication origin.
FIG. 3
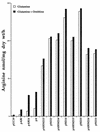
Arginine biosynthesis in E. coli DH5α containing the engineered plasmids or the parent vectors. The relevant expressed genes of vectors are shown in Table 2. The induced cultures were incubated with glutamine (20 mM) or with glutamine (20 mM) plus ornithine (5 mM) for 3 h, and total arginine production was determined and reported as nanomoles of arginine per milligram (dry weight) per hour.
FIG. 4
Multiple-sequence alignment of argA homologs. The sequences used were argA homologs from E. coli (1) (arga_ecoli), Pseudomonas aeruginosa (4) (arga_psea), and Pseudomonas putida (4) (arga_psepu). The argA homologs were aligned by using the PILEUP and PRETTY programs of the Genetics Computer Group. The three prokaryotic NAGS sequences showed 45% identity and an additional 12% similarity. Identical residues are shaded, and homologous residues detected by the PAM250 matrix of amino acid similarity (3) (determined by using the SEQVU program and manual editing) are boxed (functionally similar amino acids follow: D and E; F and Y; G and W; N and D; K and R; Q and E; L and M; I and V; and A, S, and T). The mutations H15Y (argA214), Y19C (argA215), S54N (argA218), R58H (argA213), G287S (argA216), and Q432R (argA219) are indicated above the sequence. Gaps introduced to optimize alignment are indicated by hyphens.
Similar articles
- Enhanced production of arginine and urea by genetically engineered Escherichia coli K-12 strains.
Tuchman M, Rajagopal BS, McCann MT, Malamy MH. Tuchman M, et al. Appl Environ Microbiol. 1997 Jan;63(1):33-8. doi: 10.1128/aem.63.1.33-38.1997. Appl Environ Microbiol. 1997. PMID: 8979336 Free PMC article. - Primary structure, partial purification and regulation of key enzymes of the acetyl cycle of arginine biosynthesis in Bacillus stearothermophilus: dual function of ornithine acetyltransferase.
Sakanyan V, Charlier D, Legrain C, Kochikyan A, Mett I, Piérard A, Glansdorff N. Sakanyan V, et al. J Gen Microbiol. 1993 Mar;139(3):393-402. doi: 10.1099/00221287-139-3-393. J Gen Microbiol. 1993. PMID: 8473852 - Mammalian N-acetylglutamate synthase.
Morizono H, Caldovic L, Shi D, Tuchman M. Morizono H, et al. Mol Genet Metab. 2004 Apr;81 Suppl 1(Suppl 1):S4-11. doi: 10.1016/j.ymgme.2003.10.017. Mol Genet Metab. 2004. PMID: 15050968 Free PMC article. Review. - The N-Acetylglutamate Synthase Family: Structures, Function and Mechanisms.
Shi D, Allewell NM, Tuchman M. Shi D, et al. Int J Mol Sci. 2015 Jun 9;16(6):13004-22. doi: 10.3390/ijms160613004. Int J Mol Sci. 2015. PMID: 26068232 Free PMC article. Review.
Cited by
- Over-expression of a tomato N-acetyl-L-glutamate synthase gene (SlNAGS1) in Arabidopsis thaliana results in high ornithine levels and increased tolerance in salt and drought stresses.
Kalamaki MS, Alexandrou D, Lazari D, Merkouropoulos G, Fotopoulos V, Pateraki I, Aggelis A, Carrillo-López A, Rubio-Cabetas MJ, Kanellis AK. Kalamaki MS, et al. J Exp Bot. 2009;60(6):1859-71. doi: 10.1093/jxb/erp072. Epub 2009 Apr 8. J Exp Bot. 2009. PMID: 19357433 Free PMC article. - Reengineering of a Corynebacterium glutamicum L-arginine and L-citrulline producer.
Ikeda M, Mitsuhashi S, Tanaka K, Hayashi M. Ikeda M, et al. Appl Environ Microbiol. 2009 Mar;75(6):1635-41. doi: 10.1128/AEM.02027-08. Epub 2009 Jan 9. Appl Environ Microbiol. 2009. PMID: 19139237 Free PMC article. - Site-directed mutagenesis studies on the L-arginine-binding sites of feedback inhibition in N-acetyl-L-glutamate kinase (NAGK) from Corynebacterium glutamicum.
Xu M, Rao Z, Dou W, Jin J, Xu Z. Xu M, et al. Curr Microbiol. 2012 Feb;64(2):164-72. doi: 10.1007/s00284-011-0042-y. Epub 2011 Nov 19. Curr Microbiol. 2012. PMID: 22101454 - Characterization of the argA gene required for arginine biosynthesis and syringomycin production by Pseudomonas syringae pv. syringae.
Lu SE, Soule JD, Gross DC. Lu SE, et al. Appl Environ Microbiol. 2003 Dec;69(12):7273-80. doi: 10.1128/AEM.69.12.7273-7280.2003. Appl Environ Microbiol. 2003. PMID: 14660376 Free PMC article. - Inversion of allosteric effect of arginine on N-acetylglutamate synthase, a molecular marker for evolution of tetrapods.
Haskins N, Panglao M, Qu Q, Majumdar H, Cabrera-Luque J, Morizono H, Tuchman M, Caldovic L. Haskins N, et al. BMC Biochem. 2008 Sep 18;9:24. doi: 10.1186/1471-2091-9-24. BMC Biochem. 2008. PMID: 18801197 Free PMC article.
References
- Dayhoff M O, Schwarz R M, Orcutt B C. A model of evolutionary change in proteins. Matrices for detecting distant relationships. In: Dayhoff M O, editor. Atlas of protein sequence and structure, vol. 5, suppl. 3. Washington, D.C: National Biomedical Research Foundation; 1978. pp. 345–358.
- Dharmsthiti S, Krishnapillai V. DNA sequence conservation at the gene level in a conserved chromosomal segment in two Pseudomonas species. J Genet. 1993;72:1–14.
- Eckhardt T, Leisinger T. Isolation and characterization of mutants with a feedback resistant N-acetylglutamate synthase in Escherichia coli K 12. Mol Gen Genet. 1975;138:225–232. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases