Simultaneous detection and identification of human parainfluenza viruses 1, 2, and 3 from clinical samples by multiplex PCR - PubMed (original) (raw)
Simultaneous detection and identification of human parainfluenza viruses 1, 2, and 3 from clinical samples by multiplex PCR
J E Echevarría et al. J Clin Microbiol. 1998 May.
Abstract
Reverse transcription (RT)-PCR assays have been widely described for use in the diagnosis of human parainfluenza viruses (HPIVs) and other respiratory virus pathogens. However, these assays are mostly monospecific, requiring separate amplifications for each HPIV type. In the present work, we describe multiplex RT-PCR assays that detect and differentiate HPIV serotypes 1, 2, and 3 in a combined reaction. Specifically, a mixture of three pairs of primers to conserved regions of the hemagglutinin-neuraminidase gene of each HPIV serotype was used for primary amplification, yielding amplicons with similar sizes. For typing, a second amplification was performed with a mixture of nested primers, yielding amplicons with sizes easily differentiated by agarose gel electrophoresis. A modified single-amplification RT-PCR assay with fluorescence-labeled nested primers, followed by analysis of the labeled products on an automated sequencing gel, was also evaluated. Fifteen temporally and geographically diverse HPIV isolates from the Centers for Disease Control and Prevention archives and 26 of 30 (87%) previously positive nasopharyngeal specimens (8 of 10 positive for HPIV serotype 1 [HPIV1], 9 of 10 positive for HPIV2, and 9 of 10 positive for HPIV3) were positive and were correctly typed by both assays. Negative results were obtained with naso- or oropharyngeal specimens and/or culture isolates of 33 unrelated respiratory tract pathogens, including HPIV4, enterovirus, rhinovirus, respiratory syncytial virus, adenovirus, influenza virus, and Streptococcus pneumoniae. Our multiplex RT-PCR assays provide sensitive, specific, and simplified tools for the rapid diagnosis of HPIV infections.
Figures
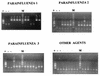
FIG. 1
Detection and typing of prototype HPIV1, HPIV2, and HPIV3 strains. (A) Lane M, molecular weight markers; lanes 1, 2, and 3, HPIV1, HPIV2, and HPIV3, respectively; lane A, mixture of all three HPIV types. (B) GeneScan Analysis Software profile of the mixture in lane A.
FIG. 2
Results of nested RT-PCR with clinical samples. No discrepancies with the results obtained by the method with GeneScan Analysis Software were found (data not shown).
Similar articles
- [Simultaneous detection of human parainfluenza viruses 1, 2, 3 by multiplex real-time reverse transcription-polymerase chain reaction with LNA probes].
Ji YX, Mao NY, Wang HH, Xie ZD, Xu WB. Ji YX, et al. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2012 Oct;26(5):388-90. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2012. PMID: 23547466 Chinese. - Detection and identification of human parainfluenza viruses 1, 2, 3, and 4 in clinical samples of pediatric patients by multiplex reverse transcription-PCR.
Aguilar JC, Pérez-Breña MP, García ML, Cruz N, Erdman DD, Echevarría JE. Aguilar JC, et al. J Clin Microbiol. 2000 Mar;38(3):1191-5. doi: 10.1128/JCM.38.3.1191-1195.2000. J Clin Microbiol. 2000. PMID: 10699020 Free PMC article. - Rapid molecular epidemiologic studies of human parainfluenza viruses based on direct sequencing of amplified DNA from a multiplex RT-PCR assay.
Echevarría JE, Erdman DD, Meissner HC, Anderson L. Echevarría JE, et al. J Virol Methods. 2000 Jul;88(1):105-9. doi: 10.1016/s0166-0934(00)00163-4. J Virol Methods. 2000. PMID: 10921847 - The landscape of extrapulmonary manifestations of human parainfluenza viruses: A systematic narrative review.
Farahmand M, Shatizadeh Malekshahi S, Jabbari MR, Shayestehpour M. Farahmand M, et al. Microbiol Immunol. 2021 Jan;65(1):1-9. doi: 10.1111/1348-0421.12865. Epub 2020 Dec 31. Microbiol Immunol. 2021. PMID: 33270253 - Parainfluenza viruses.
Henrickson KJ. Henrickson KJ. Clin Microbiol Rev. 2003 Apr;16(2):242-64. doi: 10.1128/CMR.16.2.242-264.2003. Clin Microbiol Rev. 2003. PMID: 12692097 Free PMC article. Review.
Cited by
- Real-time PCR in virology.
Mackay IM, Arden KE, Nitsche A. Mackay IM, et al. Nucleic Acids Res. 2002 Mar 15;30(6):1292-305. doi: 10.1093/nar/30.6.1292. Nucleic Acids Res. 2002. PMID: 11884626 Free PMC article. Review. - Infantile hypertrophic pyloric stenosis: are viruses involved?
Mcheik JN, Dichamp I, Levard G, Ragot S, Beby-Defaux A, Grosos C, Couvrat V, Agius G. Mcheik JN, et al. J Med Virol. 2010 Dec;82(12):2087-91. doi: 10.1002/jmv.21913. J Med Virol. 2010. PMID: 20981797 Free PMC article. - Molecular characterization of human parainfluenza virus type 1 in infants attending Mbagathi District Hospital, Nairobi, Kenya: a retrospective study.
Kiptinness JK, Wurapa EK, Wamunyokoli F, Bulimo WD. Kiptinness JK, et al. Virus Genes. 2013 Dec;47(3):439-47. doi: 10.1007/s11262-013-0970-7. Epub 2013 Aug 17. Virus Genes. 2013. PMID: 23955068 - Multiplex PCR: optimization and application in diagnostic virology.
Elnifro EM, Ashshi AM, Cooper RJ, Klapper PE. Elnifro EM, et al. Clin Microbiol Rev. 2000 Oct;13(4):559-70. doi: 10.1128/CMR.13.4.559. Clin Microbiol Rev. 2000. PMID: 11023957 Free PMC article. Review. - Poor outcome of acute respiratory infection in young children with underlying health condition in Brazil.
Durigon GS, Oliveira DB, Felicio MC, Finelli C, Pereira MF, Storni JG, Caldeira RN, Berezin RC, Durigon EL, Berezin EN. Durigon GS, et al. Int J Infect Dis. 2015 May;34:3-7. doi: 10.1016/j.ijid.2015.03.003. Epub 2015 Mar 5. Int J Infect Dis. 2015. PMID: 25747778 Free PMC article.
References
- Apalsch A M, Green M, Ledesmamedina J, Nour B, Wald E R. Parainfluenza and influenza virus infections in pediatric organ transplant recipients. Clin Infect Dis. 1995;21:394–399. - PubMed
- Arola M, Ruuskanen O, Ziegler T, Salmi T T. Respiratory virus infections during anticancer treatment in children. Pediatr Infect Dis J. 1995;14:690–694. - PubMed
- Casas I, Powell L, Klaper P E, Cleator G M. New method for the extraction of viral RNA and DNA from cerebrospinal fluid for use in the polymerase chain reaction assay. J Virol Methods. 1995;53:25–36. - PubMed
- Collins P L, Chanock R M, McIntosh K. Parainfluenza viruses. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publications; 1996. pp. 1205–1241.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources