Inactivation of a defined active site in the mouse 20S proteasome complex enhances major histocompatibility complex class I antigen presentation of a murine cytomegalovirus protein - PubMed (original) (raw)
Inactivation of a defined active site in the mouse 20S proteasome complex enhances major histocompatibility complex class I antigen presentation of a murine cytomegalovirus protein
G Schmidtke et al. J Exp Med. 1998.
Abstract
Proteasomes generate peptides bound by major histocompatibility complex (MHC) class I molecules. Avoiding proteasome inhibitors, which in most cases do not distinguish between individual active sites within the cell, we used a molecular genetic approach that allowed for the first time the in vivo analysis of defined proteasomal active sites with regard to their significance for antigen processing. Functional elimination of the delta/low molecular weight protein (LMP) 2 sites by substitution with a mutated inactive LMP2 T1A subunit results in reduced cell surface expression of the MHC class I H-2Ld and H-2Dd molecules. Surface levels of H-2Ld and H-2Dd molecules were restored by external loading with peptides. However, as a result of the active site mutation, MHC class I presentation of a 9-mer peptide derived from a protein of murine cytomegalovirus was enhanced about three- to fivefold. Our experiments provide evidence that the delta/LMP2 active site elimination limits the processing and presentation of several peptides, but may be, nonetheless, beneficial for the generation and presentation of others.
Figures
Figure 1
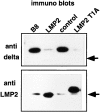
Expression of either LMP2 or the mutant LMP2 T1A results in complete replacement of subunit δ in 20S proteasomes. 20S proteasomes purified from the cell line B8 and from LMP2–, control–, and LMP2 T1A–transfected cells were examined for the presence of subunit δ (top) and LMP2 (bottom) by Western blotting. Even after strong overexposure of the immunoblots, only negligible amounts of residual δ subunit can be identified in 20S proteasomes of B8-LMP2 and B8-LMP2 T1A cells. Arrows, the position of a 21-kD marker protein.
Figure 2
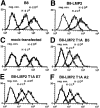
Active site mutation results in decreased surface levels of H-2Ld and H-2Dd alleles. Analysis of surface expression of the MHC class I molecules H-2Ld and H-2Dd by flow cytometry. Logarithmic fluorescence is plotted versus cell count. neg. con., the second stage control only. Shown are the histograms of the stainings of B8 (A), of the LMP2 transfectant (B), and of B8 transfected with control vector (C). Also, several different clones of the LMP2 T1A transfectants were tested; the presented data of the clones LMP2 T1A B5 (D), LMP2 T1A E7 (E), and LMP2 T1A A2 (F) show a 40% reduction of H-2Ld and H-2Dd expression. Identical results were obtained in all independent experiments performed with two different anti–H-2Ld and anti–H-2Dd antibodies.
Figure 3
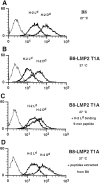
Reconstitution of surface expression of MHC class I on B8-LMP2 T1A cells by externally added peptides. Analysis of surface expression of the MHC class I molecules H-2Ld and H-2Dd by flow cytometry. Logarithmic fluorescence is plotted versus cell count. Curves drawn by thin lines represent the second stage control. Cells were cultured 18 h at 27°C in the presence of acid extracted peptides or a synthetic 9-mer known to bind H-2Ld. (A) Histograms of B8 cells (control) incubation with peptides does not alter MHC expression. (B) LMP2 T1A–transfected cells incubated in the absence of external peptides. (C) B8-LMP2 T1A cells incubated in the presence of a synthetic 9-mer known to bind to H-2Ld molecules. H-2Ld expression increases, whereas H-2Dd is not affected. (D) The externally added peptides from B8 cells restore the surface expression of MHC class I molecules on B8-LMP2 T1A cells.
Figure 4
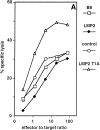
Active site mutation leads to enhanced presentation of the pp89 MCMV 9-mer peptide. (A) Analysis of MCMV pp89 antigen presentation in the cell lines B8 (white squares), LMP2 (black diamonds), control (white circles), and LMP2 T1A (white triangles). The susceptibility to lysis by MCMV pp89 (amino acids 168–176/H-2Ld) -specific cytotoxic T cells was determined in a 4-h chromium release assay. Effector/target ratios are plotted versus the percent of specific lysis. Values represent the means of three replicate cultures. Identical results were obtained in four independent experiments with two different B8-LMP2 clones and three different B8-LMP2 T1A clones. (B) Northern blot analysis of mRNA expression of the MCMV IE-I protein pp89. C4 is a BALB/c mouse-derived fibroblast cell line from which B8 was obtained by transfection with the pp89 cDNA. B8 cells were stably transfected with the LMP2 cDNA expression construct (LMP2), vector only (control), or with a construct expressing the mutated cDNA of LMP2 (LMP2 T1A). Blotted poly A+ mRNA prepared from C4, B8, B8-LMP2, control, and B8-LMP2 T1A was hybridized with a pp89 cDNA probe. All cell lines contain almost identical amounts of pp89 mRNA according to PhosphorImager analysis.
Figure 4
Active site mutation leads to enhanced presentation of the pp89 MCMV 9-mer peptide. (A) Analysis of MCMV pp89 antigen presentation in the cell lines B8 (white squares), LMP2 (black diamonds), control (white circles), and LMP2 T1A (white triangles). The susceptibility to lysis by MCMV pp89 (amino acids 168–176/H-2Ld) -specific cytotoxic T cells was determined in a 4-h chromium release assay. Effector/target ratios are plotted versus the percent of specific lysis. Values represent the means of three replicate cultures. Identical results were obtained in four independent experiments with two different B8-LMP2 clones and three different B8-LMP2 T1A clones. (B) Northern blot analysis of mRNA expression of the MCMV IE-I protein pp89. C4 is a BALB/c mouse-derived fibroblast cell line from which B8 was obtained by transfection with the pp89 cDNA. B8 cells were stably transfected with the LMP2 cDNA expression construct (LMP2), vector only (control), or with a construct expressing the mutated cDNA of LMP2 (LMP2 T1A). Blotted poly A+ mRNA prepared from C4, B8, B8-LMP2, control, and B8-LMP2 T1A was hybridized with a pp89 cDNA probe. All cell lines contain almost identical amounts of pp89 mRNA according to PhosphorImager analysis.
Similar articles
- Presentation of viral antigens restricted by H-2Kb, Db or Kd in proteasome subunit LMP2- and LMP7-deficient cells.
Zhou X, Momburg F, Liu T, Abdel Motal UM, Jondal M, Hämmerling GJ, Ljunggren HG. Zhou X, et al. Eur J Immunol. 1994 Aug;24(8):1863-8. doi: 10.1002/eji.1830240822. Eur J Immunol. 1994. PMID: 8056044 - Selective involvement of proteasomes and cysteine proteases in MHC class I antigen presentation.
López D, Del Val M. López D, et al. J Immunol. 1997 Dec 15;159(12):5769-72. J Immunol. 1997. PMID: 9550370 - Immunological functions of the proteasome.
Niedermann G. Niedermann G. Curr Top Microbiol Immunol. 2002;268:91-136. doi: 10.1007/978-3-642-59414-4_5. Curr Top Microbiol Immunol. 2002. PMID: 12083010 Review. - The function of the proteasome system in MHC class I antigen processing.
Stoltze L, Nussbaum AK, Sijts A, Emmerich NP, Kloetzel PM, Schild H. Stoltze L, et al. Immunol Today. 2000 Jul;21(7):317-9. doi: 10.1016/s0167-5699(00)01665-0. Immunol Today. 2000. PMID: 10950502 Review. No abstract available.
Cited by
- Factors regulated by interferon gamma and hypoxia-inducible factor 1A contribute to responses that protect mice from Coccidioides immitis infection.
Woelk CH, Zhang JX, Walls L, Viriyakosol S, Singhania A, Kirkland TN, Fierer J. Woelk CH, et al. BMC Microbiol. 2012 Sep 24;12:218. doi: 10.1186/1471-2180-12-218. BMC Microbiol. 2012. PMID: 23006927 Free PMC article. - Efficient generation of a hepatitis B virus cytotoxic T lymphocyte epitope requires the structural features of immunoproteasomes.
Sijts AJ, Ruppert T, Rehermann B, Schmidt M, Koszinowski U, Kloetzel PM. Sijts AJ, et al. J Exp Med. 2000 Feb 7;191(3):503-14. doi: 10.1084/jem.191.3.503. J Exp Med. 2000. PMID: 10662796 Free PMC article. - The proteasome activator 11 S REG (PA28) and class I antigen presentation.
Rechsteiner M, Realini C, Ustrell V. Rechsteiner M, et al. Biochem J. 2000 Jan 1;345 Pt 1(Pt 1):1-15. Biochem J. 2000. PMID: 10600633 Free PMC article. Review. - Repression of interferon regulatory factor 1 by hepatitis C virus core protein results in inhibition of antiviral and immunomodulatory genes.
Ciccaglione AR, Stellacci E, Marcantonio C, Muto V, Equestre M, Marsili G, Rapicetta M, Battistini A. Ciccaglione AR, et al. J Virol. 2007 Jan;81(1):202-14. doi: 10.1128/JVI.01011-06. Epub 2006 Oct 18. J Virol. 2007. PMID: 17050603 Free PMC article. - Intracellular HIV-1 Tat protein represses constitutive LMP2 transcription increasing proteasome activity by interfering with the binding of IRF-1 to STAT1.
Remoli AL, Marsili G, Perrotti E, Gallerani E, Ilari R, Nappi F, Cafaro A, Ensoli B, Gavioli R, Battistini A. Remoli AL, et al. Biochem J. 2006 Jun 1;396(2):371-80. doi: 10.1042/BJ20051570. Biochem J. 2006. PMID: 16512786 Free PMC article.
References
- Rock KL, Gramm G, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. - PubMed
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. - PubMed
- Groettrup M, Soza A, Kuckelkorn U, Kloetzel P-M. Peptide antigen production by the proteasome: complexity provides efficiency. Immunol Today. 1996;17:429–435. - PubMed
- Lehner PJ, Cresswell P. Processing and delivery of peptides presented by the MHC class I molecules. Curr Opin Immunol. 1996;8:59–67. - PubMed
- Seemüller E, Lupas A, Stock D, Löwe J, Huber R, Baumeister W. Proteasome from Thermoplasma acidophilum: a threonine protease. Science. 1995;268:579–582. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous