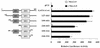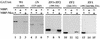GATA-4 and Nkx-2.5 coactivate Nkx-2 DNA binding targets: role for regulating early cardiac gene expression - PubMed (original) (raw)
GATA-4 and Nkx-2.5 coactivate Nkx-2 DNA binding targets: role for regulating early cardiac gene expression
J L Sepulveda et al. Mol Cell Biol. 1998 Jun.
Abstract
The cardiogenic homeodomain factor Nkx-2.5 and serum response factor (SRF) provide strong transcriptional coactivation of the cardiac alpha-actin (alphaCA) promoter in fibroblasts (C. Y. Chen and R. J. Schwartz, Mol. Cell. Biol. 16:6372-6384, 1996). We demonstrate here that Nkx-2.5 also cooperates with GATA-4, a dual C-4 zinc finger transcription factor expressed in early cardiac progenitor cells, to activate the alphaCA promoter and a minimal promoter, containing only multimerized Nkx-2.5 DNA binding sites (NKEs), in heterologous CV-1 fibroblasts. Transcriptional activity requires the N-terminal activation domain of Nkx-2.5 and Nkx-2.5 binding activity through its homeodomain but does not require GATA-4's activation domain. The minimal interactive regions were mapped to the homeodomain of Nkx-2.5 and the second zinc finger of GATA-4. Removal of Nkx-2.5's C-terminal inhibitory domain stimulated robust transcriptional activity, comparable to the effects of GATA-4 on wild-type Nkx-2.5, which in part facilitated Nkx-2.5 DNA binding activity. We postulate the following simple model: GATA-4 induces a conformational change in Nkx-2.5 that displaces the C-terminal inhibitory domain, thus eliciting transcriptional activation of promoters containing Nkx-2.5 DNA binding targets. Therefore, alphaCa promoter activity appears to be regulated through the combinatorial interactions of at least three cardiac tissue-enriched transcription factors, Nkx-2.5, GATA-4, and SRF.
Figures
FIG. 1
Synergistic activation of the αCA promoter and the NKE multimerized A20(3) promoter by Nkx-2.5 and GATA-4. (A) CV-1 cells were transfected with luciferase reporter genes (1 μg of DNA) containing the αCA promoter, the −100-bp deletion mutant of αCA containing a serum response element (αCA–Del-100), and the simian virus 40 (SV40) early promoter. Cotransfectants contained CMV promoter-directed expression vectors driving Nkx-2.5 or GATA-4 (400 μg of DNA) or the empty vector pCG (1 μg of DNA). Luciferase activity was assayed 3 days after transfection. The bars represent the averages of data from two independent transfections, and the error bars represent the standard deviations of corrected luciferase activity relative to that of the pCG vector in a typical experiment. (B) Reporter vectors contained either 58 bp of the αCA promoter upstream of the transcriptional start site (aCA-58) or the A20 Nkx-2.5 binding site cloned in triplicate upstream of aCA-58 [A20(3)]. Nkx-2.5pm is a mutant of Nkx-2.5 with greatly decreased DNA binding affinity.
FIG. 2
GATA-4 and Nkx-2.5 do not display cross-binding specificities. (A) GATA-4 does not bind NKEs. Nuclear extracts were prepared from CV-1 cells transfected with the Nkx-2.5 expression vector (pCGN–Nkx-2.5) or the empty vector (pCGN). Five micrograms of either the control extract (lane 2) or the Nkx-2.5 extract (lanes 3 to 7) was preincubated with 1 μg of poly(dG-dC) at room temperature for 15 min in 20 μl of 1× binding buffer; 50-fold molar excesses of unlabeled double-stranded specific (lane 4) and nonspecific (lanes 5 to 7) competitors were included in the reaction mixtures. Following the preincubation, 0.02 pmol of end-labeled A20 oligonucleotides was added, and the reaction mixtures were incubated for a further 15 min. DNA-protein complexes were resolved on a 5% polyacrylamide gel cast and run in 0.5× Tris-borate-EDTA. (B and C) Reaction conditions were similar to those described for panel A. One microliter of unprogrammed (lane 2) or GATA-4 RNA-programmed (lanes 3 to 8) rabbit reticulocyte lysate was used instead of nuclear extract. The GATA-4 binding site from the rat BNP promoter was the probe. The specific and nonspecific competitors used are indicated above the lanes. GATA con contains two GATA-4 consensus DNA binding sites (in boldface) (CACTTGATAACAGAAAGTGATAACTCT), and GATA mut has mutated GATA sites (underlined) (CACTT
CT
TAACAGAAAGT
CT
TAACTCT); αCA-luc is the αCA promoter-containing, pGL2-derived reporter vector described previously (11), and GATA-4-luc (G4-luc) is pGL2 containing three copies of the αC-MHC GATA binding site. Specific DNA-protein complexes are indicated by arrowheads.
FIG. 3
Coactivation with GATA-4 is a property of some, but not all, NK-2 family members. CV-1 cells were transfected, as described in the legend to Fig. 1, with expression vectors coding for Nkx-2.5, Tinman, Nkx-2.1, and Nkx-2.8 in the presence of GATA-4 (black bars) or pCG (white bars). Reporter constructs were αCA–Del-100 (A) and A20(3)-Luc (B). Relative luciferase activities were assayed and reported as described in the legend to Fig. 1.
FIG. 4
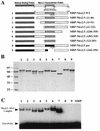
Mapping of Nkx-2.5 domains required for transcriptional synergism with GATA-4. CV-1 cells were transfected with A20(3)-Luc, various mutants of Nkx-2.5, and either GATA-4 (black bars) or pCG (white bars). The numbers represent the amino acid range conserved or deleted (▵) in each mutant. The diagrams on the left represent the various conserved domains of Nkx-2.5 retained on each mutant. TN, tinman domain; +++, the positively charged N-terminal activation domain; homeo, homeobox; NK, conserved hydrophobic NK-2 motif. Luciferase activity was assayed as described in the legend to Fig. 1. Note the different scales of corrected luciferase activity for the two panels.
FIG. 5
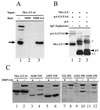
The N-terminal and C-terminal domains inhibit Nkx-2.5 homeodomain DNA binding activity. (A) Schematic diagrams of MBP–Nkx-2.5 deletion mutants; (B) MBP–Nkx-2.5 proteins purified on amylose beads, separated by 10% PAGE, and detected by Coomassie blue staining; (C) results of an EMSA using equivalent amounts of purified MBP–Nkx-2.5 proteins and a labeled A20 oligonucleotide. EMSA conditions were as described in the legend to Fig. 2. Lane numbers in panels B and C correspond to the fusion proteins represented in panel A.
FIG. 6
GATA-4 facilitates DNA binding of full-length Nkx-2.5 but not the homeodomain. (A) End-labeled double-stranded A20 oligonucleotide probe was incubated with 10 or 25 ng of MBP–Nkx-2.5 homeodomain (HD), either with 250 ng of MBP–GATA-4 (lanes 3 and 7, respectively) or without MBP–GATA-4 (lanes 2 and 5, respectively), in 1× binding buffer. Lanes 4 and 6 contained only MBP–GATA-4 and A20 probe. The faint bands in lane 1 are due to spillover from lane 2. (B) MBP–Nkx-2.5 (10 or 25 ng of purified protein) was incubated with (lanes 7 and 8, respectively) or without (lanes 3 and 4, respectively) 250 ng of MBP–GATA-4 for 15 min at 30°C in 1× binding buffer. End-labeled double-stranded NKE (0.02 pmol) was added to the protein mixtures, which were subsequently incubated for an additional 15 min. Under identical conditions, 10 ng (lane 1), 25 ng (lane 2), or 250 ng (lane 5) of MBP and 250 ng of MBP–GATA-4 (lane 6) did not bind the labeled NKE probe. DNA-protein complexes are indicated by arrowheads. The gel in panel B was exposed overnight, and the one in panel A was exposed for 4 days at −70°C.
FIG. 7
Pull-down protein association assays with MBP–GATA-4 and HA-tagged Nkx-2.5 protein extracts from transfected CV-1 cells. (A) MBP protein (lane 2) or MBP–GATA-4 fusion protein (MBP-G4) (lane 3) was purified from bacterial extracts, immobilized on amylose beads, and mixed with whole-cell extracts of CV-1 cells transfected with HA epitope-tagged wild-type Nkx-2.5 (Nkx-2.5 wt). After being washed, bound proteins were identified by immunoblotting. Lane 1 represents 25% of the input cellular extract. The arrow points to the Nkx-2.5-specific band. Other bands represent nonspecific cross-reactivity of the antibody. (B) Whole-cell extracts of CV-1 cells cotransfected with pCGN–Nkx-2.5 and expression vectors for either protein A (pA) (lane 3) or protein A–GATA-4 (pA-GATA4) (lane 2) were incubated with IgG-Sepharose beads and washed extensively. Immobilized protein complexes were eluted by boiling in SDS sample buffer and visualized by immunoblotting with anti-HA antibody. Note that proteins containing the protein A domains (arrowheads), as well as IgG heavy (γ) and light (κ,λ) chains, were visualized due to affinity for the secondary antibody. Lane 1 represents 25% of the input extract from pCGN–Nkx-2.5- and protein–GATA-4 transfected cells. (C) Immobilized MBP–GATA-4 protein (MBP-G4) purified from bacterial extract was mixed with whole-cell extract of CV-1 cells transfected with either HA epitope-tagged wild-type Nkx-2.5 (Nkx-2.5 wt) or various Nkx-2.5 mutants (see Fig. 3 for diagrams). After being washed, bound proteins were separated by SDS-PAGE (lanes 1, 3, 5, 7, 9, and 11). Lanes 2, 4, 6, 8, 10, and 12 represent 20% of the input cellular extract. Immunoblotting was performed with anti-HA antibody. Specific bands representing Nkx-2.5 mutants are identified by a broken-line box.
FIG. 8
Mapping GATA-4 domains required for transcriptional synergism with Nkx-2.5. CV-1 fibroblasts were transfected with A20(3)-Luc, various mutants of GATA-4, and either Nkx-2.5 (black bars) or pCG (white bars). The diagrams on the left represent the domains of GATA-4 retained in each deletion mutant. ZF1 and ZF2 refer to the amino- and carboxy-terminal zinc fingers of GATA-4, respectively. Luciferase activity was assayed as described in the legend to Fig. 1. wt, wild type.
FIG. 9
Physical interaction between Nkx-2.5 and GATA-4 is mediated by the C-terminal zinc finger (ZF2) of GATA-4. Ten microliters of reticulocyte lysate containing in vitro-translated, 35S-labeled wild-type (Wt) GATA-4 (lanes 1 to 3), an N-terminally truncated GATA-4 mutant (ΔN) (lanes 4 to 6), GATA-4 with both zinc fingers (lanes 7 to 9), GATA-4 with only zinc finger 2 (lanes 10 to 12), or GATA-4 with only zinc finger 1 (ZF1) (lanes 13 to 15) was incubated with approximately 10 μg of MBP (lanes 2, 5, 8, 11, and 14) or MBP–Nkx-2.5 (lanes 3, 6, 9, 12, and 15). The beads were washed extensively, and the bound proteins were resolved on SDS–10% polyacrylamide gels and visualized by autoradiography. Lanes 1, 4, 7, 10, and 13 contained 10% of the in vitro translation volume used for binding reactions. After being washed, bound proteins were visualized by fluorography.
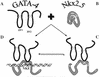
FIG. 10
Working model for activation of Nkx-2.5 by interaction with GATA-4. GATA-4 interacts with Nkx-2.5 and induces a conformational change resulting in increased affinity for the Nkx-2.5 binding site (NKE) and increased accessibility of the activation domain (+++). (A) Isolated GATA-4 protein; (B) isolated Nkx-2.5 protein. The model postulates that in the inactivated state the Nkx-2.5 homeodomain (HD) has a low affinity for NKE due to steric hindrance by the C-terminal domain, possibly via hydrophobic interactions with the N terminus of Nkx-2.5. In addition, it is possible that the C-terminal–N-terminal interactions keep the activation domain inaccessible to the transcriptional machinery. (C) Interaction with GATA-4 would then remove this negative effect of the C terminus on Nkx-2.5 transcriptional activity. (D) NKE binding by activated Nkx-2.5–GATA-4 complexes. It is also possible that GATA-4 leaves the complex before or after NKE binding (data not shown). ZF1 and ZF2, zinc fingers 1 and 2, respectively.
Similar articles
- Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac alpha-actin gene transcription.
Chen CY, Schwartz RJ. Chen CY, et al. Mol Cell Biol. 1996 Nov;16(11):6372-84. doi: 10.1128/MCB.16.11.6372. Mol Cell Biol. 1996. PMID: 8887666 Free PMC article. - Activation of the cardiac alpha-actin promoter depends upon serum response factor, Tinman homologue, Nkx-2.5, and intact serum response elements.
Chen CY, Croissant J, Majesky M, Topouzis S, McQuinn T, Frankovsky MJ, Schwartz RJ. Chen CY, et al. Dev Genet. 1996;19(2):119-30. doi: 10.1002/(SICI)1520-6408(1996)19:2<119::AID-DVG3>3.0.CO;2-C. Dev Genet. 1996. PMID: 8900044 - Context-dependent transcriptional cooperation mediated by cardiac transcription factors Csx/Nkx-2.5 and GATA-4.
Shiojima I, Komuro I, Oka T, Hiroi Y, Mizuno T, Takimoto E, Monzen K, Aikawa R, Akazawa H, Yamazaki T, Kudoh S, Yazaki Y. Shiojima I, et al. J Biol Chem. 1999 Mar 19;274(12):8231-9. doi: 10.1074/jbc.274.12.8231. J Biol Chem. 1999. PMID: 10075728 - GATA transcription factors and cardiac development.
Charron F, Nemer M. Charron F, et al. Semin Cell Dev Biol. 1999 Feb;10(1):85-91. doi: 10.1006/scdb.1998.0281. Semin Cell Dev Biol. 1999. PMID: 10355032 Review. - Regulation of heart development and function through combinatorial interactions of transcription factors.
Nemer G, Nemer M. Nemer G, et al. Ann Med. 2001 Dec;33(9):604-10. doi: 10.3109/07853890109002106. Ann Med. 2001. PMID: 11817655 Review.
Cited by
- Cardiac tissue inhibitor of matrix metalloprotease 4 dictates cardiomyocyte contractility and differentiation of embryonic stem cells into cardiomyocytes: Road to therapy.
Chaturvedi P, Kalani A, Familtseva A, Kamat PK, Metreveli N, Tyagi SC. Chaturvedi P, et al. Int J Cardiol. 2015 Apr 1;184:350-363. doi: 10.1016/j.ijcard.2015.01.091. Epub 2015 Feb 25. Int J Cardiol. 2015. PMID: 25745981 Free PMC article. - Effect of Rehmannia glutinosa Libosch extract on proliferation and cardiogenic pre-differentiation of human mesenchymal stem cells.
Nguyen HD, Ho Thi L, Ho XB, Cao VA, Le Hoang DM. Nguyen HD, et al. Biomedicine (Taipei). 2022 Mar 1;12(1):39-52. doi: 10.37796/2211-8039.1243. eCollection 2022. Biomedicine (Taipei). 2022. PMID: 35836917 Free PMC article. - The Drosophila Transcription Factors Tinman and Pannier Activate and Collaborate with Myocyte Enhancer Factor-2 to Promote Heart Cell Fate.
Lovato TL, Sensibaugh CA, Swingle KL, Martinez MM, Cripps RM. Lovato TL, et al. PLoS One. 2015 Jul 30;10(7):e0132965. doi: 10.1371/journal.pone.0132965. eCollection 2015. PLoS One. 2015. PMID: 26225919 Free PMC article. - Forward Programming of Cardiac Stem Cells by Homogeneous Transduction with MYOCD plus TBX5.
Belian E, Noseda M, Abreu Paiva MS, Leja T, Sampson R, Schneider MD. Belian E, et al. PLoS One. 2015 Jun 5;10(6):e0125384. doi: 10.1371/journal.pone.0125384. eCollection 2015. PLoS One. 2015. PMID: 26047103 Free PMC article. - Intramolecular control of transcriptional activity by the NK2-specific domain in NK-2 homeodomain proteins.
Watada H, Mirmira RG, Kalamaras J, German MS. Watada H, et al. Proc Natl Acad Sci U S A. 2000 Aug 15;97(17):9443-8. doi: 10.1073/pnas.97.17.9443. Proc Natl Acad Sci U S A. 2000. PMID: 10944215 Free PMC article.
References
- Azpiazu N, Frasch M. tinman and bagpipe: two homeobox genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993;7:1325–1340. - PubMed
- Bellaguli N, Schildmeyer L, Schwartz R J. Organization and myogenic restricted expression of the murine serum response factor gene: a role for autoregulation. J Biol Chem. 1997;272:18222–18231. - PubMed
- Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118:719–729. - PubMed
- Bohinski R J, Di Lauro R, Whitsett J A. The lung-specific surfactant protein B gene promoter is a target for thyroid transcription factor 1 and hepatocyte nuclear factor 3, indicating common factors for organ-specific gene expression along the foregut axis. Mol Cell Biol. 1994;14:5671–5681. - PMC - PubMed
- Brazas R M, Bhoite L T, Murphy M D, Yu Y, Chen Y, Neklason D W, Stillman D J. Determining the requirements for cooperative DNA binding by Swi5p and Pho2p (Grf10p/Bas2p) at the HO promoter. J Biol Chem. 1995;270:29151–29161. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous