Epithelial V-like antigen (EVA), a novel member of the immunoglobulin superfamily, expressed in embryonic epithelia with a potential role as homotypic adhesion molecule in thymus histogenesis - PubMed (original) (raw)
Epithelial V-like antigen (EVA), a novel member of the immunoglobulin superfamily, expressed in embryonic epithelia with a potential role as homotypic adhesion molecule in thymus histogenesis
M Guttinger et al. J Cell Biol. 1998.
Abstract
Thymus development depends on a complex series of interactions between thymocytes and the stromal component of the organ. To identify regulated genes during this codependent developmental relationship, we have applied an RNA fingerprinting technique to the analysis of thymus expansion and maturation induced in recombinase-deficient mice injected with anti-CD3 antibodies. This approach led us to the identification of a gene encoding a new member of the immunoglobulin superfamily, named epithelial V-like antigen (EVA), which is expressed in thymus epithelium and strongly downregulated by thymocyte developmental progression. This gene is expressed in the thymus and in several epithelial structures early in embryogenesis. EVA is highly homologous to the myelin protein zero and, in thymus-derived epithelial cell lines, is poorly soluble in nonionic detergents, strongly suggesting an association to the cytoskeleton. Its capacity to mediate cell adhesion through a homophilic interaction and its selective regulation by T cell maturation might imply the participation of EVA in the earliest phases of thymus organogenesis.
Figures
Figure 1
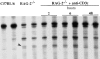
RNA fingerprinting experiment comparing cDNAs of thymi from C57BL/6, untreated RAG-2−/− mice and RAG-2−/− mice at different times (2, 6, and 48 h) after i.v. injection of anti-CD3ε mAb. RNA fingerprinting RT-PCR reactions were conducted in duplicate at each stage examined. Arrowhead; band corresponding to the Eva transcript.
Figure 2
(A) Alignment of the aa sequences encoded by human and murine Eva cDNAs. Left, aa numbering. The predicted leader (signal peptide) and transmembrane sequences are labeled together with the following features: dark-shaded boxes, potential N-linked glycosylation sites; light-shaded boxes, cysteine residues (47 and 123) found in conserved positions within the V-type domain; bold residues, potential protein casein kinase 2 phosphorylation site. These sequence data are available under GenBank/ EMBL/DDBJ accession numbers AF030455 (for human Eva) and AF030454 (for mouse Eva). (B) Model of EVA protein according to prediction results. \\, N-linked glycosylation sites; P, putative protein CK2 phosphorylation site in the cytoplasmic tail. (C) Sequence alignment of mouse EVA and myelin Po proteins. Boxed residues are identical in the two proteins (33%); homologous residues are shaded (45%).
Figure 3
Mapping of Eva in the mouse genome. Top: haplotype and linkage analysis of Eva and flanking loci on mouse chromosome 9 through the analysis of the BSS backcross (The Jackson Laboratory). Empty squares, Mus spretus allele; solid squares, C57BL/6J allele; stippled squares, genotype not determined. Numbers to the right, between rows, indicate recombination fractions ± standard error, and LOD scores. Columns represent different haplotypes observed on chromosome 9. Numbers below columns define the number of individuals sharing each haplotype. Bottom: position of Eva on chromosome 9 with respect to nearby markers independently mapped by others on the BSS backcross. Numbers on the left represent approximate genetic distances (cM) from the most centromeric chromosome 9 marker in this cross. Chromosomal locations of three syntenic human genes, including EVA, are also indicated.
Figure 4
(A) Eva mRNA expression in wild-type and RAG-2−/− thymi. Northern analysis on total RNA isolated from thymi of C57BL/6, untreated RAG-2−/− mice and RAG-2−/− mice at different times (2, 6, and 48 h) after anti-CD3ε mAb injection. After hybridization with a labeled cDNA specific for Eva, the filter was stripped and again hybridized with a labeled β-actin probe. The apparent size of the transcript is indicated. (B) Eva mRNA expression in various adult mouse tissues. RNase protection assay with a 437-bp _Eva_-specific antisense riboprobe on seven adult mouse tissues and thymi of C57BL/6, untreated RAG-2−/− mice, and RAG-2−/− mice i.v. injected with anti-CD3ε mAb. Results obtained with mouse β-actin riboprobe are also shown. (C) Eva mRNA expression in different thymic epithelial cell lines. RT-PCR on mRNA of four thymic epithelial cell lines (A2T, A89A, TEC, and TNC.R3.1). Gene expression on RAG-2−/− thymus was used as control. cDNA-amplified fragment is 412-bp long.
Figure 4
(A) Eva mRNA expression in wild-type and RAG-2−/− thymi. Northern analysis on total RNA isolated from thymi of C57BL/6, untreated RAG-2−/− mice and RAG-2−/− mice at different times (2, 6, and 48 h) after anti-CD3ε mAb injection. After hybridization with a labeled cDNA specific for Eva, the filter was stripped and again hybridized with a labeled β-actin probe. The apparent size of the transcript is indicated. (B) Eva mRNA expression in various adult mouse tissues. RNase protection assay with a 437-bp _Eva_-specific antisense riboprobe on seven adult mouse tissues and thymi of C57BL/6, untreated RAG-2−/− mice, and RAG-2−/− mice i.v. injected with anti-CD3ε mAb. Results obtained with mouse β-actin riboprobe are also shown. (C) Eva mRNA expression in different thymic epithelial cell lines. RT-PCR on mRNA of four thymic epithelial cell lines (A2T, A89A, TEC, and TNC.R3.1). Gene expression on RAG-2−/− thymus was used as control. cDNA-amplified fragment is 412-bp long.
Figure 4
(A) Eva mRNA expression in wild-type and RAG-2−/− thymi. Northern analysis on total RNA isolated from thymi of C57BL/6, untreated RAG-2−/− mice and RAG-2−/− mice at different times (2, 6, and 48 h) after anti-CD3ε mAb injection. After hybridization with a labeled cDNA specific for Eva, the filter was stripped and again hybridized with a labeled β-actin probe. The apparent size of the transcript is indicated. (B) Eva mRNA expression in various adult mouse tissues. RNase protection assay with a 437-bp _Eva_-specific antisense riboprobe on seven adult mouse tissues and thymi of C57BL/6, untreated RAG-2−/− mice, and RAG-2−/− mice i.v. injected with anti-CD3ε mAb. Results obtained with mouse β-actin riboprobe are also shown. (C) Eva mRNA expression in different thymic epithelial cell lines. RT-PCR on mRNA of four thymic epithelial cell lines (A2T, A89A, TEC, and TNC.R3.1). Gene expression on RAG-2−/− thymus was used as control. cDNA-amplified fragment is 412-bp long.
Figure 5
mRNA in situ hybridization of RAG-2−/− thymi. Eva expression by RAG-2−/−thymi from untreated mice (A and B) and treated animals at 48 h after injection of antibody (C and D). Bright-field views (A and C) and dark-field images (B and D). Note strong, homogeneous labeling of RAG-2−/− thymus, and absence of Eva expression in the thymus from injected mice.
Figure 6
Eva mRNA in situ hybridization at 13.5 d p.c. (A and B) and 14.5 d p.c. (C and D) embryonic tissue sections. Bright-field images (A and C) and dark-field microphotographs (B and D). (A and B) High levels of Eva transcripts are found in the emerging thymus (A, arrows). Specific labeling can also be seen in epithelia lining the respiratory and gastrointestinal tracts: trachea, esophagus, gut and alveoli are indicated by arrowheads. (C and D) Note labeling of surface ectoderm and submandibulary salivary glands (arrowheads). Bars: (A and B), 165 μm; (C and D), 330 μm.
Figure 7
(A) In vitro translation of mouse Eva transcript. In vitro translation was performed in the absence (lanes 1, 3, and 5) or in the presence (lanes 2, 4, and 6) of canine pancreatic microsomal membranes. Translated polypeptides were fractionated through a 12% SDS-PAGE gel and then analyzed by fluorography. Lanes 1 and 2: no RNA as negative control. Lanes 3 and 4: Saccharomyces cerevisiae α-factor mRNA, used as positive control for both translation and glycosylation. The α-factor and the core-glycosylated precursor have molecular weights of 18.6 and 32 kD, respectively. Lanes 5 and 6, Eva mRNA. Lanes 7 and 8: Eva mRNA as in lane 6 or treated with the enzyme endo-H. Positions of molecular mass markers (in kDs) are indicated on the right. (B) Cellular localization and immunoblot of chimeric EVA. Left, anti-myc immunofluorescence on CHO cells transfected with either pcDNA.3 vector (top) or Eva.myc vector (bottom). Right, anti-myc immunoblot on Triton X-100 lysates from the same cells. (C) Anti-EVA immunoblot. Western analysis with rabbit anti-EVA serum of Triton X-100 lysates from CHO cells transfected with either pcDNA.3 vector or Eva.myc vector and lysates in SDS of thymic epithelial cell lines A89A and TNC.R3.1. (D) EVA recovery in insoluble cell fraction. TNC.R3.1 and A89A cells were lysed in hypotonic buffer and separated into three fractions: P1, pellet of low-speed centrifugation; P100, pellet recovered after 100,000 g centrifugation, S100, soluble fraction. Equivalent amounts of all samples were resolved by SDS-PAGE in a 5–15% gradient gel and immunoblotted with rabbit anti-EVA antiserum, anti-actin and anti-src mAbs. EVA is exclusively present in the P1 fraction of TNC.R3.1 cells.
Figure 8
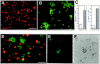
EVA-mediated cell aggregation. Nonrecombinant pcDNA.3-transfected CHO and Eva.myc-transfected CHO were detached and labeled, either with PKH26 or PKH2, respectively. After dissociation, the two cell lines were allowed to aggregate in suspension culture, either alone or mixed in a 1:1 ratio, and then analyzed 90 min after incubation in low Ca buffer. Vector-transfected cells are scattered as a single cell suspension (A), whereas large cell clusters are formed by Eva.myc- expressing cells (B). Extent of aggregation is expressed by aggregation index, calculated as D = (N0 - N90)/N0. Large aggregates (>5 cells) represented the 8.5% of the total events in mock-transfected cells and 60% in Eva-myc cells (C). In the coculture, EVA-expressing cells aggregate on their own; random incorporation of parental CHO into aggregates never exceeded 10–20% (D). Coaggregation experiments with unlabeled cells were done as described above. Immunofluorescence staining with anti-myc mAb shows selective labeling of cells forming aggregates (E), when compared with phase-contrast analysis of the same field (F). Bars: (A and B) 50 μm; (D–F) 25 μm.
Similar articles
- EVA regulates thymic stromal organisation and early thymocyte development.
DeMonte L, Porcellini S, Tafi E, Sheridan J, Gordon J, Depreter M, Blair N, Panigada M, Sanvito F, Merati B, Albientz A, Barthlott T, Ozmen L, Blackburn CC, Guttinger M. DeMonte L, et al. Biochem Biophys Res Commun. 2007 May 4;356(2):334-40. doi: 10.1016/j.bbrc.2007.02.131. Epub 2007 Mar 5. Biochem Biophys Res Commun. 2007. PMID: 17362876 - Cloning of an immunoglobulin family adhesion molecule selectively expressed by endothelial cells.
Hirata Ki, Ishida T, Penta K, Rezaee M, Yang E, Wohlgemuth J, Quertermous T. Hirata Ki, et al. J Biol Chem. 2001 May 11;276(19):16223-31. doi: 10.1074/jbc.M100630200. Epub 2001 Feb 13. J Biol Chem. 2001. PMID: 11279107 - CLMP, a novel member of the CTX family and a new component of epithelial tight junctions.
Raschperger E, Engstrom U, Pettersson RF, Fuxe J. Raschperger E, et al. J Biol Chem. 2004 Jan 2;279(1):796-804. doi: 10.1074/jbc.M308249200. Epub 2003 Oct 22. J Biol Chem. 2004. PMID: 14573622 - The murine homolog of human Ep-CAM, a homotypic adhesion molecule, is expressed by thymocytes and thymic epithelial cells.
Nelson AJ, Dunn RJ, Peach R, Aruffo A, Farr AG. Nelson AJ, et al. Eur J Immunol. 1996 Feb;26(2):401-8. doi: 10.1002/eji.1830260220. Eur J Immunol. 1996. PMID: 8617310 - Bves, a member of the Popeye domain-containing gene family.
Osler ME, Smith TK, Bader DM. Osler ME, et al. Dev Dyn. 2006 Mar;235(3):586-93. doi: 10.1002/dvdy.20688. Dev Dyn. 2006. PMID: 16444674 Free PMC article. Review.
Cited by
- Inactivation of the RB family prevents thymus involution and promotes thymic function by direct control of Foxn1 expression.
Garfin PM, Min D, Bryson JL, Serwold T, Edris B, Blackburn CC, Richie ER, Weinberg KI, Manley NR, Sage J, Viatour P. Garfin PM, et al. J Exp Med. 2013 Jun 3;210(6):1087-97. doi: 10.1084/jem.20121716. Epub 2013 May 13. J Exp Med. 2013. PMID: 23669396 Free PMC article. - MPZL2, Encoding the Epithelial Junctional Protein Myelin Protein Zero-like 2, Is Essential for Hearing in Man and Mouse.
Wesdorp M, Murillo-Cuesta S, Peters T, Celaya AM, Oonk A, Schraders M, Oostrik J, Gomez-Rosas E, Beynon AJ, Hartel BP, Okkersen K, Koenen HJPM, Weeda J, Lelieveld S, Voermans NC, Joosten I, Hoyng CB, Lichtner P, Kunst HPM, Feenstra I, de Bruijn SE; DOOFNL Consortium; Admiraal RJC, Yntema HG, van Wijk E, Del Castillo I, Serra P, Varela-Nieto I, Pennings RJE, Kremer H. Wesdorp M, et al. Am J Hum Genet. 2018 Jul 5;103(1):74-88. doi: 10.1016/j.ajhg.2018.05.011. Epub 2018 Jun 28. Am J Hum Genet. 2018. PMID: 29961571 Free PMC article. - A monoclonal antibody against the extracellular domain of mouse and human epithelial V-like antigen 1 reveals a restricted expression pattern among CD4- CD8- thymocytes.
Garabatos N, Blanco J, Fandos C, Lopez E, Santamaria P, Ruiz A, Perez-Vidakovics ML, Benveniste P, Galkin O, Zuñiga-Pflucker JC, Serra P. Garabatos N, et al. Monoclon Antib Immunodiagn Immunother. 2014 Oct;33(5):305-11. doi: 10.1089/mab.2014.0030. Monoclon Antib Immunodiagn Immunother. 2014. PMID: 25357997 Free PMC article. - Gene expression profiling for the purposes of biomarker discovery in oral potentially malignant lesions: a systematic review.
Abdulmajeed AA, Farah CS. Abdulmajeed AA, et al. Clin Med Insights Oncol. 2013 Oct 31;7:279-90. doi: 10.4137/CMO.S12950. Clin Med Insights Oncol. 2013. PMID: 24250244 Free PMC article. Review. - Large-scale screening for novel low-affinity extracellular protein interactions.
Bushell KM, Söllner C, Schuster-Boeckler B, Bateman A, Wright GJ. Bushell KM, et al. Genome Res. 2008 Apr;18(4):622-30. doi: 10.1101/gr.7187808. Epub 2008 Feb 22. Genome Res. 2008. PMID: 18296487 Free PMC article.
References
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Auerbach R. Morphogenetic interactions in the development of the mouse thymus gland. Dev Biol. 1960;2:271–285. - PubMed
- Austyn JM, Gordon S. F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur J Immunol. 1981;11:805–815. - PubMed
- Ausubel, F.M., R. Brent, R.E. Kingstone, D.D. Moore, J.A. Smith, and K.R. Struhl. 1995. Current Protocols in Molecular Biology. John Wiley and Sons, New York.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases