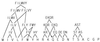Highly specific protein sequence motifs for genome analysis - PubMed (original) (raw)
Highly specific protein sequence motifs for genome analysis
C G Nevill-Manning et al. Proc Natl Acad Sci U S A. 1998.
Abstract
We present a method for discovering conserved sequence motifs from families of aligned protein sequences. The method has been implemented as a computer program called EMOTIF (http://motif. stanford.edu/emotif). Given an aligned set of protein sequences, EMOTIF generates a set of motifs with a wide range of specificities and sensitivities. EMOTIF also can generate motifs that describe possible subfamilies of a protein superfamily. A disjunction of such motifs often can represent the entire superfamily with high specificity and sensitivity. We have used EMOTIF to generate sets of motifs from all 7,000 protein alignments in the BLOCKS and PRINTS databases. The resulting database, called IDENTIFY (http://motif. stanford.edu/identify), contains more than 50,000 motifs. For each alignment, the database contains several motifs having a probability of matching a false positive that range from 10(-10) to 10(-5). Highly specific motifs are well suited for searching entire proteomes, while generating very few false predictions. IDENTIFY assigns biological functions to 25-30% of all proteins encoded by the Saccharomyces cerevisiae genome and by several bacterial genomes. In particular, IDENTIFY assigned functions to 172 of proteins of unknown function in the yeast genome.
Figures
Figure 1
Substitution groups. Groups of amino acids found to occur together in columns of aligned sequences in both the
blocks
and
hssp
databases. Only groups of amino acids that occur together at a significant frequency and are separated from all other amino acids at a level of significance of less than 0.01 are included. The substitution groups are arranged hierarchically to show relationships between their physical properties.
Figure 2
Aligned block of 34 tubulin proteins and two motifs representing these sequences. (a) An aligned block of 34 tubulin proteins and the sequence variation observed among them. (b) One possible sequence motif for the alignment in a that can be formed by using the amino acid substitution groups from Fig. 1. (c) A much more specific sequence motif that can be used to represent the upper 19 tubulin sequences, which form a group more closely related to each other than to the lower 15 sequences.
Figure 3
Enumeration of tubulin motifs by
emotif
.
emotif
generates all possible sequence motifs that can cover at least 30% of 159 tubulin sequences in a training set. Each motif is plotted as a dot in the figure where the horizontal axis gives the coverage of the motif (number of sequences covered in the training set), and the vertical axis plots the specificity of the motif as the probability of matching a random protein segment. The motifs occur in vertical lines because coverage is an integer quantity. The lower curve is the Pareto-optimal curve, which represents the most specific motif at each level of sensitivity.
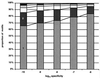
Figure 4
The number of motifs required to cover at least 90% of the protein family in the
identify
database.
emotif
was used to generate one or more motifs that cover at least 90% of all the sequences in each of 7,000 alignments in the
blocks
or
prints
databases at five different levels of specificity. Plotted are the number of motifs that are required to cover at least 90% of the sequences in the alignment.
Similar articles
- The EMOTIF database.
Huang JY, Brutlag DL. Huang JY, et al. Nucleic Acids Res. 2001 Jan 1;29(1):202-4. doi: 10.1093/nar/29.1.202. Nucleic Acids Res. 2001. PMID: 11125091 Free PMC article. - eBLOCKs: enumerating conserved protein blocks to achieve maximal sensitivity and specificity.
Su QJ, Lu L, Saxonov S, Brutlag DL. Su QJ, et al. Nucleic Acids Res. 2005 Jan 1;33(Database issue):D178-82. doi: 10.1093/nar/gki060. Nucleic Acids Res. 2005. PMID: 15608172 Free PMC article. - Saccharomyces Genome Database (SGD) provides biochemical and structural information for budding yeast proteins.
Weng S, Dong Q, Balakrishnan R, Christie K, Costanzo M, Dolinski K, Dwight SS, Engel S, Fisk DG, Hong E, Issel-Tarver L, Sethuraman A, Theesfeld C, Andrada R, Binkley G, Lane C, Schroeder M, Botstein D, Michael Cherry J. Weng S, et al. Nucleic Acids Res. 2003 Jan 1;31(1):216-8. doi: 10.1093/nar/gkg054. Nucleic Acids Res. 2003. PMID: 12519985 Free PMC article. - Beyond complete genomes: from sequence to structure and function.
Koonin EV, Tatusov RL, Galperin MY. Koonin EV, et al. Curr Opin Struct Biol. 1998 Jun;8(3):355-63. doi: 10.1016/s0959-440x(98)80070-5. Curr Opin Struct Biol. 1998. PMID: 9666332 Review. - Protein sequence motifs.
Bork P, Koonin EV. Bork P, et al. Curr Opin Struct Biol. 1996 Jun;6(3):366-76. doi: 10.1016/s0959-440x(96)80057-1. Curr Opin Struct Biol. 1996. PMID: 8804823 Review.
Cited by
- Using amino acid physicochemical distance transformation for fast protein remote homology detection.
Liu B, Wang X, Chen Q, Dong Q, Lan X. Liu B, et al. PLoS One. 2012;7(9):e46633. doi: 10.1371/journal.pone.0046633. Epub 2012 Sep 28. PLoS One. 2012. PMID: 23029559 Free PMC article. - Using structural motif templates to identify proteins with DNA binding function.
Jones S, Barker JA, Nobeli I, Thornton JM. Jones S, et al. Nucleic Acids Res. 2003 Jun 1;31(11):2811-23. doi: 10.1093/nar/gkg386. Nucleic Acids Res. 2003. PMID: 12771208 Free PMC article. - The InterPro database, an integrated documentation resource for protein families, domains and functional sites.
Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, Durbin R, Falquet L, Fleischmann W, Gouzy J, Hermjakob H, Hulo N, Jonassen I, Kahn D, Kanapin A, Karavidopoulou Y, Lopez R, Marx B, Mulder NJ, Oinn TM, Pagni M, Servant F, Sigrist CJ, Zdobnov EM. Apweiler R, et al. Nucleic Acids Res. 2001 Jan 1;29(1):37-40. doi: 10.1093/nar/29.1.37. Nucleic Acids Res. 2001. PMID: 11125043 Free PMC article. - The EMOTIF database.
Huang JY, Brutlag DL. Huang JY, et al. Nucleic Acids Res. 2001 Jan 1;29(1):202-4. doi: 10.1093/nar/29.1.202. Nucleic Acids Res. 2001. PMID: 11125091 Free PMC article. - Finding important sites in protein sequences.
Bickel PJ, Kechris KJ, Spector PC, Wedemayer GJ, Glazer AN. Bickel PJ, et al. Proc Natl Acad Sci U S A. 2002 Nov 12;99(23):14764-71. doi: 10.1073/pnas.222508899. Epub 2002 Nov 4. Proc Natl Acad Sci U S A. 2002. PMID: 12417758 Free PMC article.
References
- Scharf M, Schneider R, Casari G, Bork P, Valencia A, Ouzounis C, Sander C. ISMB. 1994;2:348–353. - PubMed
- Casari G, Ouzounis C, Valencia A, Sander C. GeneQuiz II: Automatic Function Assignment for Genome Sequence Analysis, Pacific Symposium and Biocomputing, 1996. Kohala Coast, HI: World Scientific; 1996. pp. 707–709.
- Sonnhammer E L, Eddy S R, Durbin R. Proteins. 1997;28:405–420. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases