Genomic evidence for two functionally distinct gene classes - PubMed (original) (raw)
Genomic evidence for two functionally distinct gene classes
M C Rivera et al. Proc Natl Acad Sci U S A. 1998.
Abstract
Analyses of complete genomes indicate that a massive prokaryotic gene transfer (or transfers) preceded the formation of the eukaryotic cell. In comparisons of the entire set of Methanococcus jannaschii genes with their orthologs from Escherichia coli, Synechocystis 6803, and the yeast Saccharomyces cerevisiae, it is shown that prokaryotic genomes consist of two different groups of genes. The deeper, diverging informational lineage codes for genes which function in translation, transcription, and replication, and also includes GTPases, vacuolar ATPase homologs, and most tRNA synthetases. The more recently diverging operational lineage codes for amino acid synthesis, the biosynthesis of cofactors, the cell envelope, energy metabolism, intermediary metabolism, fatty acid and phospholipid biosynthesis, nucleotide biosynthesis, and regulatory functions. In eukaryotes, the informational genes are most closely related to those of Methanococcus, whereas the majority of operational genes are most closely related to those of Escherichia, but some are closest to Methanococcus or to Synechocystis.
Figures
Figure 1
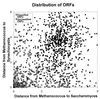
The distribution of ORFs indicates that classified ORFs are distributed differently than unclassified ORFs. This scatterplot displays the distance between a methanogen gene and its cyanobacterial ortholog on the vertical axis and the distance between the methanogen gene and its yeast ortholog on the horizontal axis. Orthologs of classified ORFs (□) are closely related (corresponding to small distances) and therefore are distributed about the origin at the lower left. In contrast, orthologs of unclassified ORFs (•) are distantly related and are distributed about (3.5, 3.8). Distance estimates between orthologous genes were obtained from
blastp
(17) probabilities (see Methods).
Figure 2
A three-dimensional display of gene orthologs classified by function or by lineage. (A) The sets of gene orthologs are labeled by their functional classes. In the stereo view (B), the classes are combined into two superclasses corresponding to whether the genes function in informational (blue) or operational (red) processes. The axes are similarity scores between the methanogen and cyanobacterial orthologs, Smc, between the methanogen and yeast orthologs, Smy, and between the proteobacterial and the cyanobacterial orthologs, Spc. The functional categories are: amino acid synthesis (A), biosynthesis of cofactors (B), cell envelope proteins (C), energy metabolism (E), GTPases and homologs of vacuolar ATPases (G), intermediary metabolism (I), fatty acid and phospholipid biosynthesis (L), nucleotide biosynthesis (N), other (O), cell processes (P), replication (R), transcription (S), translation (T), transport (X), tRNA synthetases (Y), and regulatory genes (Z). The divisions into functional categories are good, but exceptions exist such as valyl-tRNA synthetase, which appears in the operational group.
Figure 3
The prokaryotic origins of eukaryotic genes. The results of phylogenetic analyses are shown plotted with respect to similarity scores, in the orientation used in Fig. 2, using three methods of analysis; Jukes–Cantor distances (A); maximum parsimony (B); and paralinear (LogDet) distances (C). In this representation, orthologs from the informational lineage are located on the lower right cube face and orthologs from the operational lineage are located either on the upper or the lower left cube face. Trees in which the eukaryotic gene is most closely related to the Methanococcus, the Synechocystis, or the Escherichia ortholog are indicated by blue, green, and red squares, respectively.
Figure 4
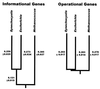
Phylogenetic trees reconstructed from gene orthologs from the informational lineage and from the operational lineage. Distances on the trees refer to paralinear (LogDet) distances in nucleotide substitutions per replacement position. The error estimates correspond to one SD measured from 100 bootstrap replicates. The trees are both rooted in the methanogen branch by using paralogus genes as described in Methods.
Figure 5
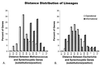
The distribution of pairwise paralinear (LogDet) distances between orthologous ORFs for informational (gray) and operational (white) genes. (A) The distributions of distances between Methanococcus and Synechocystis are significantly different for informational and operational genes, whereas in B, the distributions of distances between Escherichia and Synechocystis are similar.
Figure 6
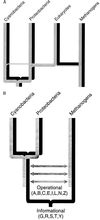
Evolution of the operational and informational lineages. The, complex, evolution of eukaryotes is shown in A. The informational lineage (black) is inherited entirely from a methanogen ancestor, whereas the operational lineage (gray) is inherited principally from a proteobacterial ancestor, although methanogen and cyanobacterial ancestors also make a significant contribution. The prokaryotic evolution of the operational (gray) and informational (black) lineages is shown in B. Horizontal arrows indicate possible lateral gene transfers (see text).
Similar articles
- Comparison of archaeal and bacterial genomes: computer analysis of protein sequences predicts novel functions and suggests a chimeric origin for the archaea.
Koonin EV, Mushegian AR, Galperin MY, Walker DR. Koonin EV, et al. Mol Microbiol. 1997 Aug;25(4):619-37. doi: 10.1046/j.1365-2958.1997.4821861.x. Mol Microbiol. 1997. PMID: 9379893 - Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics.
Smith DR, Doucette-Stamm LA, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Reeve JN, et al. Smith DR, et al. J Bacteriol. 1997 Nov;179(22):7135-55. doi: 10.1128/jb.179.22.7135-7155.1997. J Bacteriol. 1997. PMID: 9371463 Free PMC article. - Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii.
Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, Kerlavage AR, Dougherty BA, Tomb JF, Adams MD, Reich CI, Overbeek R, Kirkness EF, Weinstock KG, Merrick JM, Glodek A, Scott JL, Geoghagen NS, Venter JC. Bult CJ, et al. Science. 1996 Aug 23;273(5278):1058-73. doi: 10.1126/science.273.5278.1058. Science. 1996. PMID: 8688087 - Reconstruction of amino acid biosynthesis pathways from the complete genome sequence.
Bono H, Ogata H, Goto S, Kanehisa M. Bono H, et al. Genome Res. 1998 Mar;8(3):203-10. doi: 10.1101/gr.8.3.203. Genome Res. 1998. PMID: 9521924 Review. - A genomic perspective on protein families.
Tatusov RL, Koonin EV, Lipman DJ. Tatusov RL, et al. Science. 1997 Oct 24;278(5338):631-7. doi: 10.1126/science.278.5338.631. Science. 1997. PMID: 9381173 Review.
Cited by
- A universal and constant rate of gene content change traces pangenome flux to LUCA.
Trost K, Knopp MR, Wimmer JLE, Tria FDK, Martin WF. Trost K, et al. FEMS Microbiol Lett. 2024 Jan 9;371:fnae068. doi: 10.1093/femsle/fnae068. FEMS Microbiol Lett. 2024. PMID: 39165128 Free PMC article. - Chapter 5: Major Biological Innovations in the History of Life on Earth.
Bozdag GO, Szeinbaum N, Conlin PL, Chen K, Fos SM, Garcia A, Penev PI, Schaible GA, Trubl G. Bozdag GO, et al. Astrobiology. 2024 Mar;24(S1):S107-S123. doi: 10.1089/ast.2021.0119. Astrobiology. 2024. PMID: 38498818 - Interaction with a phage gene underlie costs of a β-lactamase.
Lai H-Y, Cooper TF. Lai H-Y, et al. mBio. 2024 Feb 14;15(2):e0277623. doi: 10.1128/mbio.02776-23. Epub 2024 Jan 9. mBio. 2024. PMID: 38194254 Free PMC article. - Origins and Functional Significance of Eukaryotic Protein Folds.
Romei M, Carpentier M, Chomilier J, Lecointre G. Romei M, et al. J Mol Evol. 2023 Dec;91(6):854-864. doi: 10.1007/s00239-023-10136-x. Epub 2023 Dec 7. J Mol Evol. 2023. PMID: 38060007 - Genome-wide transformation reveals extensive exchange across closely related Bacillus species.
Förster M, Rathmann I, Yüksel M, Power JJ, Maier B. Förster M, et al. Nucleic Acids Res. 2023 Dec 11;51(22):12352-12366. doi: 10.1093/nar/gkad1074. Nucleic Acids Res. 2023. PMID: 37971327 Free PMC article.
References
- Sogin M L. Curr Opin Genet Dev. 1991;1:457–463. - PubMed
- Golding G B, Gupta R S. Mol Biol Evol. 1994;12:1–6. - PubMed
- Doolittle W F. In: Evolution of Microbial Life. Roberts D M, Sharp P, Alderson G, Collins M A, editors. Cambridge, U.K.: Cambridge Univ. Press; 1996. pp. 1–21.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases