t(11;22)(q23;q11.2) In acute myeloid leukemia of infant twins fuses MLL with hCDCrel, a cell division cycle gene in the genomic region of deletion in DiGeorge and velocardiofacial syndromes - PubMed (original) (raw)
Case Reports
t(11;22)(q23;q11.2) In acute myeloid leukemia of infant twins fuses MLL with hCDCrel, a cell division cycle gene in the genomic region of deletion in DiGeorge and velocardiofacial syndromes
M D Megonigal et al. Proc Natl Acad Sci U S A. 1998.
Erratum in
- Proc Natl Acad Sci U S A 1998 Aug 18;95(17):10344
Abstract
We examined the MLL genomic translocation breakpoint in acute myeloid leukemia of infant twins. Southern blot analysis in both cases showed two identical MLL gene rearrangements indicating chromosomal translocation. The rearrangements were detectable in the second twin before signs of clinical disease and the intensity relative to the normal fragment indicated that the translocation was not constitutional. Fluorescence in situ hybridization with an MLL-specific probe and karyotype analyses suggested t(11;22)(q23;q11. 2) disrupting MLL. Known 5' sequence from MLL but unknown 3' sequence from chromosome band 22q11.2 formed the breakpoint junction on the der(11) chromosome. We used panhandle variant PCR to clone the translocation breakpoint. By ligating a single-stranded oligonucleotide that was homologous to known 5' MLL genomic sequence to the 5' ends of BamHI-digested DNA through a bridging oligonucleotide, we formed the stem-loop template for panhandle variant PCR which yielded products of 3.9 kb. The MLL genomic breakpoint was in intron 7. The sequence of the partner DNA from band 22q11.2 was identical to the hCDCrel (human cell division cycle related) gene that maps to the region commonly deleted in DiGeorge and velocardiofacial syndromes. Both MLL and hCDCrel contained homologous CT, TTTGTG, and GAA sequences within a few base pairs of their respective breakpoints, which may have been important in uniting these two genes by translocation. Reverse transcriptase-PCR amplified an in-frame fusion of MLL exon 7 to hCDCrel exon 3, indicating that an MLL-hCDCrel chimeric mRNA had been transcribed. Panhandle variant PCR is a powerful strategy for cloning translocation breakpoints where the partner gene is undetermined. This application of the method identified a region of chromosome band 22q11.2 involved in both leukemia and a constitutional disorder.
Figures
Figure 1
Schematic of template generation and amplification of genomic translocation breakpoint on a der(11) chromosome by panhandle variant PCR.
Figure 2
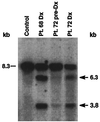
Identification of rearrangements within the MLL bcr by Southern blot in leukemias of infant twins 68 and 72 and in peripheral blood mononuclear cells (PBMCs) of twin 72 7 weeks before the onset of clinical leukemia. Intensity of identical rearrangements in preleukemic PBMCs of twin 72 relative to the germ-line band suggests that the translocation was not constitutional. _Bam_HI-digested DNAs were hybridized with B859 human MLL cDNA probe spanning exons 5–11 (13). The 8.3-kb fragment is from the unrearranged allele (dash marks). Control DNA from PBMCs of a normal individual shows the germ-line pattern. Arrows show rearrangements.
Figure 3
(A) Panhandle variant PCR products from der (11) chromosome of t(11;22)(q23;q11.2) in AMLs of infant twins 68 and 72. (B) Summary of t(11;22) breakpoint junction and partner DNA identified by directly sequencing panhandle variant PCR products from AML of patient 68 and sequencing products of panhandle variant PCR from leukemias of patients 68 and 72 subcloned by recombination PCR. Comparison with normal sequence identified the breakpoint at nucleotide 2,672 in MLL intron 7 (bold arrow). Primer 3 serves as the primer in the final PCR (see Fig. 1). The 5′ 2,622 bp include primer 3 and MLL bcr sequence to breakpoint at position 2,672 in intron 7. The 1,240 bp of 3′ sequence are partner DNA, and the most 3′ 31 bp of sequence are the complement of the ligated oligonucleotide (i.e., the complement of primer 3). Underlines show short segments of homology between MLL and hCDC_rel_ at respective breakpoint junctions. Positions of relevant cosmid clones in chromosome band 22q11.2 deletion syndromes are shown at top (GenBank accession nos. AC000093 and AC000091). rc indicates that GenBank entry is reverse complement of hCDC_rel_.
Figure 4
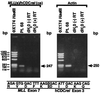
RT–PCR analysis of total RNA from leukemic cells of patient 68 indicating MLL_-hCDC_rel chimeric mRNA. RT–PCRs with sense and antisense primers from MLL exon 6 and hCDC_rel_ exon 3, respectively, and randomly primed cDNA template gave a single 247-bp product (Upper). Direct sequencing of the products of four reactions revealed an in-frame fusion of MLL exon 7 to hCDC_rel_ exon 3 at position 142 of the 2,032-bp full-length hCDC_rel_ cDNA (GenBank accession no. U74628) (16) (Lower). Reactions using β-actin primers and RNA negative control (distilled H2O) are also shown (Upper).
Similar articles
- MLL-SEPTIN6 fusion recurs in novel translocation of chromosomes 3, X, and 11 in infant acute myelomonocytic leukaemia and in t(X;11) in infant acute myeloid leukaemia, and MLL genomic breakpoint in complex MLL-SEPTIN6 rearrangement is a DNA topoisomerase II cleavage site.
Slater DJ, Hilgenfeld E, Rappaport EF, Shah N, Meek RG, Williams WR, Lovett BD, Osheroff N, Autar RS, Ried T, Felix CA. Slater DJ, et al. Oncogene. 2002 Jul 11;21(30):4706-14. doi: 10.1038/sj.onc.1205572. Oncogene. 2002. PMID: 12096348 - Panhandle PCR strategy to amplify MLL genomic breakpoints in treatment-related leukemias.
Megonigal MD, Rappaport EF, Jones DH, Kim CS, Nowell PC, Lange BJ, Felix CA. Megonigal MD, et al. Proc Natl Acad Sci U S A. 1997 Oct 14;94(21):11583-8. doi: 10.1073/pnas.94.21.11583. Proc Natl Acad Sci U S A. 1997. PMID: 9326653 Free PMC article. - Panhandle polymerase chain reaction amplifies MLL genomic translocation breakpoint involving unknown partner gene.
Felix CA, Kim CS, Megonigal MD, Slater DJ, Jones DH, Spinner NB, Stump T, Hosler MR, Nowell PC, Lange BJ, Rappaport EF. Felix CA, et al. Blood. 1997 Dec 15;90(12):4679-86. Blood. 1997. PMID: 9389682 - MLL-SEPT5 fusion transcript in infant acute myeloid leukemia with t(11;22)(q23;q11).
Launay E, Henry C, Meyer C, Chappé C, Taque S, Boulland ML, Ben Abdelali R, Dugay F, Marschalek R, Bastard C, Fest T, Gandemer V, Belaud-Rotureau MA. Launay E, et al. Leuk Lymphoma. 2014 Mar;55(3):662-7. doi: 10.3109/10428194.2013.809528. Epub 2013 Aug 20. Leuk Lymphoma. 2014. PMID: 23725386 Review. - Rearrangements involving chromosome band 11Q23 in acute leukaemia.
Rowley JD. Rowley JD. Semin Cancer Biol. 1993 Dec;4(6):377-85. Semin Cancer Biol. 1993. PMID: 8142623 Review.
Cited by
- The mammalian septin MSF localizes with microtubules and is required for completion of cytokinesis.
Surka MC, Tsang CW, Trimble WS. Surka MC, et al. Mol Biol Cell. 2002 Oct;13(10):3532-45. doi: 10.1091/mbc.e02-01-0042. Mol Biol Cell. 2002. PMID: 12388755 Free PMC article. - Subcellular localization of EEN/endophilin A2, a fusion partner gene in leukaemia.
Cheung N, So CW, Yam JW, So CK, Poon RY, Jin DY, Chan LC. Cheung N, et al. Biochem J. 2004 Oct 1;383(Pt 1):27-35. doi: 10.1042/BJ20040041. Biochem J. 2004. PMID: 15214844 Free PMC article. - MSF (MLL septin-like fusion), a fusion partner gene of MLL, in a therapy-related acute myeloid leukemia with a t(11;17)(q23;q25).
Osaka M, Rowley JD, Zeleznik-Le NJ. Osaka M, et al. Proc Natl Acad Sci U S A. 1999 May 25;96(11):6428-33. doi: 10.1073/pnas.96.11.6428. Proc Natl Acad Sci U S A. 1999. PMID: 10339604 Free PMC article. - A Bioinformatic Pipeline Places STAT5A as a miR-650 Target in Poorly Differentiated Aggressive Breast Cancer.
López-Huerta E, Fuentes-Pananá EM. López-Huerta E, et al. Int J Mol Sci. 2020 Oct 19;21(20):7720. doi: 10.3390/ijms21207720. Int J Mol Sci. 2020. PMID: 33086498 Free PMC article. - Identification of a gene at 11q23 encoding a guanine nucleotide exchange factor: evidence for its fusion with MLL in acute myeloid leukemia.
Kourlas PJ, Strout MP, Becknell B, Veronese ML, Croce CM, Theil KS, Krahe R, Ruutu T, Knuutila S, Bloomfield CD, Caligiuri MA. Kourlas PJ, et al. Proc Natl Acad Sci U S A. 2000 Feb 29;97(5):2145-50. doi: 10.1073/pnas.040569197. Proc Natl Acad Sci U S A. 2000. PMID: 10681437 Free PMC article.
References
- Gurney J G, Severson R K, Davis S, Robison L L. Cancer. 1995;75:2186–2195. - PubMed
- CDC. Morb Mortal Wkly Rep. 1997;46:121–125. - PubMed
- Clarkson B D, Boyse E A. Lancet. 1971;1:699–701. - PubMed
- Zueler W W, Cox D E. Semin Hematol. 1969;6:228–249. - PubMed
- Buckley J, Buckley C, Breslow N, Draper G, Roberson P, Mack T. Med Pediatr Oncol. 1996;26:223–229. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous