Restriction-modification gene complexes as selfish gene entities: roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion - PubMed (original) (raw)
Restriction-modification gene complexes as selfish gene entities: roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion
Y Nakayama et al. Proc Natl Acad Sci U S A. 1998.
Abstract
We have reported some type II restriction-modification (RM) gene complexes on plasmids resist displacement by an incompatible plasmid through postsegregational host killing. Such selfish behavior may have contributed to the spread and maintenance of RM systems. Here we analyze the role of regulatory genes (C), often found linked to RM gene complexes, in their interaction with the host and the other RM gene complexes. We identified the C gene of EcoRV as a positive regulator of restriction. A C mutation eliminated postsegregational killing by EcoRV. The C system has been proposed to allow establishment of RM systems in new hosts by delaying the appearance of restriction activity. Consistent with this proposal, bacteria preexpressing ecoRVC were transformed at a reduced efficiency by plasmids carrying the EcoRV RM gene complex. Cells carrying the BamHI RM gene complex were transformed at a reduced efficiency by a plasmid carrying a PvuII RM gene complex, which shares the same C specificity. The reduction most likely was caused by chromosome cleavage at unmodified PvuII sites by prematurely expressed PvuII restriction enzyme. Therefore, association of the C genes of the same specificity with RM gene complexes of different sequence specificities can confer on a resident RM gene complex the capacity to abort establishment of a second, incoming RM gene complex. This phenomenon, termed "apoptotic mutual exclusion," is reminiscent of suicidal defense against virus infection programmed by other selfish elements. pvuIIC and bamHIC genes define one incompatibility group of exclusion whereas ecoRVC gene defines another.
Figures
Figure 1
(A) Genetic and restriction map of _Eco_RV RMC genes (modified from ref. 27). (B) Effects of the ecoRVC mutation on the restriction and modification activities. The ratio of the number of λ plaques on the test strain to that on DH5 (pBR322) is shown (Left). The λ phage grown in each strain was assayed on DH5 (pYNEC113) and DH5 (pBR322); the ratio of the number of plaques on these two strains is shown (Right). Measurements were carried out in duplicate in two separate experiments.
Figure 2
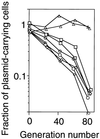
Apparent stability of plasmids with the _Eco_RV RM gene complex and its mutant versions. During repeated batch culture (106-fold dilution and overnight incubation) in the absence of any selective antibiotics, aliquots were assayed on LB agar plates with and without Ap. The ratio of these colony counts was taken as the fraction of cells carrying the plasmid. The generation number was estimated from the colony counts on LB agar. □, pBR322; ▵, pYNEC113 (R+ M+ C+); ○, pYNEC117 (_R_- M+ C+); and ◊, pYNEC118 (R+ M+ _C_−).
Figure 3
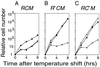
Postsegregational host cell killing following induced loss of the _Eco_RV RM gene complex. The replication of repts plasmids carrying _Eco_RV RM gene complex was blocked by a temperature shift. The bacterial cells [JC8679 (pYNEC114), JC8679 (pYNEC119), and JC8679 (pYNEC120)] were incubated at 30°C in LB broth with Ap until the OD660 of the culture reached 0.3. Antibiotics were then removed, and the temperature was shifted to 42°C. The culture was diluted when the OD660 reached 0.3 (≈5 × 108 cell/ml). The number of total cells (▪) was counted under a microscope. The number of viable cells (○) and plasmid-carrying cells (▴) were measured by counting colonies on LB agar plates and LB agar plates containing Ap, respectively.
Figure 4
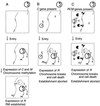
Hypotheses for the establishment and apoptotic mutual exclusion of RM gene complexes containing a C gene. (A) A plasmid carrying an RM gene complex enters a cell. M and C genes are expressed first. After the modification enzyme has modified almost all the chromosomal sites, the accumulation of C protein induces the restriction enzyme. (B) Entry of a plasmid carrying an RM gene complex into a cell carrying C gene from the same RM gene complex. The resident C protein induces the expression of the incoming R gene, whose product cleaves the unmodified chromosomal sites and kills the cell. (C) Entry of a plasmid carrying an RM gene complex into a cell harboring another RM gene complex with a different sequence specificity but with the same C protein specificity. The resident C protein induces the incoming R gene, whose product cleaves the unmodified chromosomal sites and kills the cell.
Figure 5
Effect of a resident ecoRVC on the establishment of an incoming _Eco_RV RM system. Cells carrying pYNEC126 (ecoRVC+) or pYNEC130 (ecoRVC −) were transformed with 150 ng of pYNEC113 (R+ M+ C+), pYNEC117 (_R_− M+ C+), pYNEC118 (R+ M+ _C_−), or pBR322 (vector). Each DNA solution contained 100 ng of pKC31 (KmR) as an internal control of the transformation efficiency. For each assay, the number of ApR transformants was divided by the number of KmR transformants. The relative transformation efficiencies are the above ratios further divided by the ratio for pBR322. The measurements were carried out in duplicate in two separate experiments.
Figure 6
Effect of a resident _Bam_HI RM system on the establishment of an incoming _Pvu_II RM system. Cells carrying pYNEC403 (_Bam_HI R+ M+ C+), pYNEC404 (_Bam_HI R − M+ C+) or pBR322 were transformed with 90 ng of pYNEC323 (_Pvu_II R+ M+ C+), pYNEC324 (_Pvu_II _R_− M+ C+), pYNEC325 (_Pvu_II R+M+ _C_−), or pYNEC333 (vector). Each DNA solution contained 60 ng of pYNEC332 (KmR) as an internal control of the transformation efficiency. For each assay, the number of ApR transformants was divided by the number of KmR transformants. The relative transformation efficiencies are the above ratios further divided by the ratio for pYNEC333. The measurements were carried out in duplicate in two separate experiments.
Figure 7
Effect of various C genes on the establishment of an incoming _Pvu_II RM system. Cells carrying pYNEC126 (ecoRVC+), pYNEC130 (_ecoRVC_−), pYNEC311 (pvuIIC+), pYNEC401 (bamHIC+), or pIK153 (vector) were transformed with 150 ng of pYNEC300 (_Pvu_II R+ M+ C+) or pYNEC302 (_Pvu_II _R_− M+ C+). Each DNA solution contained 100 ng of pKC31 (KmR) as an internal control of the transformation efficiency. For each assay, the number of ApR transformants was divided by the number of KmR transformants. The relative transformation efficiencies are the above ratios further divided by the ratio for pIK153. The measurements were carried out in duplicate in two separate experiments.
Similar articles
- Dependence of post-segregational killing mediated by Type II restriction-modification systems on the lifetime of restriction endonuclease effective activity.
Kozlova S, Morozova N, Ispolatov Y, Severinov K. Kozlova S, et al. mBio. 2024 Aug 14;15(8):e0140824. doi: 10.1128/mbio.01408-24. Epub 2024 Jul 9. mBio. 2024. PMID: 38980007 Free PMC article. - Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution.
Kobayashi I. Kobayashi I. Nucleic Acids Res. 2001 Sep 15;29(18):3742-56. doi: 10.1093/nar/29.18.3742. Nucleic Acids Res. 2001. PMID: 11557807 Free PMC article. Review. - Translational independence between overlapping genes for a restriction endonuclease and its transcriptional regulator.
Kaw MK, Blumenthal RM. Kaw MK, et al. BMC Mol Biol. 2010 Nov 19;11:87. doi: 10.1186/1471-2199-11-87. BMC Mol Biol. 2010. PMID: 21092102 Free PMC article. - Restriction-modification systems as genomic parasites in competition for specific sequences.
Kusano K, Naito T, Handa N, Kobayashi I. Kusano K, et al. Proc Natl Acad Sci U S A. 1995 Nov 21;92(24):11095-9. doi: 10.1073/pnas.92.24.11095. Proc Natl Acad Sci U S A. 1995. PMID: 7479944 Free PMC article. - Selfishness and death: raison d'être of restriction, recombination and mitochondria.
Kobayashi I. Kobayashi I. Trends Genet. 1998 Sep;14(9):368-74. doi: 10.1016/s0168-9525(98)01532-7. Trends Genet. 1998. PMID: 9769733 Review.
Cited by
- Regulatory circuit based on autogenous activation-repression: roles of C-boxes and spacer sequences in control of the PvuII restriction-modification system.
Mruk I, Rajesh P, Blumenthal RM. Mruk I, et al. Nucleic Acids Res. 2007;35(20):6935-52. doi: 10.1093/nar/gkm837. Epub 2007 Oct 11. Nucleic Acids Res. 2007. PMID: 17933763 Free PMC article. - Cell death upon epigenetic genome methylation: a novel function of methyl-specific deoxyribonucleases.
Fukuda E, Kaminska KH, Bujnicki JM, Kobayashi I. Fukuda E, et al. Genome Biol. 2008;9(11):R163. doi: 10.1186/gb-2008-9-11-r163. Epub 2008 Nov 21. Genome Biol. 2008. PMID: 19025584 Free PMC article. - Diverse functions of restriction-modification systems in addition to cellular defense.
Vasu K, Nagaraja V. Vasu K, et al. Microbiol Mol Biol Rev. 2013 Mar;77(1):53-72. doi: 10.1128/MMBR.00044-12. Microbiol Mol Biol Rev. 2013. PMID: 23471617 Free PMC article. Review. - Gain and loss of multiple genes during the evolution of Helicobacter pylori.
Gressmann H, Linz B, Ghai R, Pleissner KP, Schlapbach R, Yamaoka Y, Kraft C, Suerbaum S, Meyer TF, Achtman M. Gressmann H, et al. PLoS Genet. 2005 Oct;1(4):e43. doi: 10.1371/journal.pgen.0010043. PLoS Genet. 2005. PMID: 16217547 Free PMC article. - Nature of the promoter activated by C.PvuII, an unusual regulatory protein conserved among restriction-modification systems.
Knowle D, Lintner RE, Touma YM, Blumenthal RM. Knowle D, et al. J Bacteriol. 2005 Jan;187(2):488-97. doi: 10.1128/JB.187.2.488-497.2005. J Bacteriol. 2005. PMID: 15629920 Free PMC article.
References
- Kobayashi I. In: Epigenetic Mechanisms of Gene Regulation. Russo V, Martienssen R, Riggs A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 155–172.
- Naito T, Kusano K, Kobayashi I. Science. 1995;267:897–899. - PubMed
- Naito, Y., Naito, T. & Kobayashi, I. (1998) Biol. Chem. 379, in press. - PubMed
- Jensen R B, Gerdes K. Mol Microbiol. 1995;17:205–210. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases