Two nearly identical aromatic compound hydrolase genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1 - PubMed (original) (raw)
Two nearly identical aromatic compound hydrolase genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1
A Yamada et al. Appl Environ Microbiol. 1998 Jun.
Abstract
The two 2-hydroxy-6-oxohepta-2,4-dienoate (HOHD) hydrolase genes, etbD1 and etbD2, were cloned from a strong polychlorinated biphenyl (PCB) degrader, Rhodococcus sp. strain RHA1, and their nucleotide sequences were determined. The etbD2 gene was located in the vicinity of bphA gene homologs and encoded an enzyme whose amino-terminal sequence was very similar to the amino-terminal sequence of the HOHD hydrolase which was purified from RHA1. Using the etbD2 gene fragment as a probe, we cloned the etbD1 gene encoding the purified HOHD hydrolase by colony hybridization. Both genes encode a product having 274 amino acid residues and containing the nucleophile motif conserved in alpha/beta hydrolase fold enzymes. The deduced amino acid sequences were quite similar to the amino acid sequences of the products of the single-ring aromatic hydrolase genes, such as dmpD, cumD, todF, and xylF, and not very similar to the amino acid sequences of the products of bphD genes from PCB degraders, including RHA1. The two HOHD hydrolase genes and the RHA1 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate (HPDA) hydrolase gene, bphD, were expressed in Escherichia coli, and their relative enzymatic activities were examined. The product of bphD was very specific to HPDA, and the products of etbD1 and etbD2 were specific to HOHD. All of the gene products exhibited poor activities against the meta-cleavage product of catechol. These results agreed with the results obtained for BphD and EtbD1 hydrolases purified from RHA1. The three hydrolase genes exhibited similar induction patterns both in an RNA slot blot hybridization analysis and in a reporter gene assay when a promoter probe vector was used. They were induced by biphenyl, ethylbenzene, benzene, toluene, and ortho-xylene. Strain RCD1, an RHA1 mutant strain lacking both the bphD gene and the etbD2 gene, grew well on ethylbenzene. This result suggested that the etbD1 gene product is involved in the meta-cleavage metabolic pathway of ethylbenzene.
Figures
FIG. 1
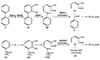
Proposed metabolic pathway for aerobic degradation of biphenyl and ethylbenzene in Rhodococcus sp. strain RHA1. Compound I, biphenyl; compound II, 23DHBP; compound III, HPDA; compound IV, 2-hydroxypenta-2,4-dienoate; compound V, benzoic acid; compound VI, ethylbenzene (toluene); compound VII, 3-ethylcatechol (3-methylcatechol); compound VIII, HOHD; compound IX, propanoic acid (acetic acid). TCA, tricarboxylic acid.
FIG. 2
Gene organizations of the DNA fragments containing the bphD (A), etbD1 (B), and etbD2 (C) hydrolase genes. Coding regions of the genes or ORFs possibly involved in aromatic compound metabolism are indicated by broad arrows. The solid bars below the gene organizations represent the fragments used to construct subclones of each hydrolase gene, whose designations are indicated on the right. The transcriptional direction of each subclone from the lac promoter of the vector plasmid is indicated by a thin arrow. Double-headed arrows indicate the fragments used for hybridization experiments. The shaded boxes represent the fragments used to construct reporter plasmids, whose designations are given below the boxes. Abbreviations: A, _Apa_I; B, _Bam_HI; E, _Eco_RI; H, _Hin_dIII; N, _Nar_I; S, _Sac_I; Sc, _Sca_I; Sp, _Sph_I; St, _Stu_I; X, _Xho_I.
FIG. 3
Nucleotide and deduced amino acid sequences of the etbD1 (A) and etbD2 (B) genes of Rhodococcus sp. strain RHA1 and their products. Putative ribosome binding sites and nucleophile motifs of α/β hydrolase fold enzymes are enclosed in boxes and underlined, respectively. Stop codons are indicated by dots.
FIG. 4
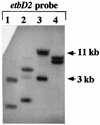
Southern hybridization analysis of genomic DNA from Rhodococcus sp. strain RHA1 performed with the etbD2 gene probe. The etbD2 gene probe used is shown in Fig. 2 (probe 3). Genomic DNA was digested with _Sac_I, _Sal_I, _Eco_RI, and _Bam_HI (lanes 1 to 4, respectively).
FIG. 5
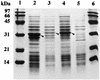
Expression of the bphD, etbD1, and etbD2 genes in E. coli. Putative gene products are indicated by arrowheads. Cell extracts of E. coli transformants grown in the presence of isopropyl-β-
d
-thiogalactopyranoside were subjected to SDS–15% polyacrylamide gel electrophoresis. Lanes 1 and 6, molecular mass markers, including phosphorylase (97 kDa), bovine serum albumin (66 kDa), ovalbumin (45 kDa), carbonic anhydrase (31 kDa), soybean trypsin inhibitor (21 kDa), and lysozyme (14 kDa); lane 2, E. coli JM109(pUAD1 carrying bphD); lane 3, E. coli JM109(pUAD2 carrying etbD1); lane 4, E. coli JM109(pUAD3 carrying etbD2); lane 5, E. coli JM109(pUC18).
FIG. 6
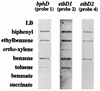
RNA slot blot hybridization analysis of _bphD_-, _etbD1_-, and _etbD2_-specific transcripts in Rhodococcus sp. strain RHA1. Two micrograms of each total RNA from RHA1 cells grown on the substrates indicated was blotted onto a nylon membrane and hybridized with DIG-labeled probes 1, 2, and 4 (Fig. 2) specific to bphD, etbD1, and etbD2, respectively.
FIG. 7
(A) Physical map of the promoter probe vector, pKLA1. (B) Luciferase activities of Rhodococcus sp. strain RHA1 harboring reporter plasmid derivatives. pKLABD1, pKLAED1, and pKLAED2 contain the promoter regions of bphD, etbD1, and etbD2, respectively, which were inserted at the _Sal_I site preceding the luxAB reporter genes of the promoter probe vector, pKLA1. Data are averages from triplicate determinations in at least three independent experiments; error bars are shown. The luciferase activities of RHA1 cells harboring pKLA1 in 0.2× LB, 0.2× LB containing biphenyl, and 0.2× LB supplemented with ethylbenzene were less than 0.1 × 106 light units per _A_600 unit. Inducer compound abbreviations: Suc, succinate; BA, benzoate; BP, biphenyl; Ben, benzene; Tol, toluene; ETB, ethylbenzene; Xyl, _ortho_-xylene. LU, light units.
FIG. 8
Southern hybridization analysis of mutant strains defective for growth on biphenyl. Genomic DNAs from RHA1, RCD1, and RCAD1 were digested with _Eco_RI and hybridized with the bphD gene probe (A) (probe 1 in Fig. 2) and the etbD2 gene probe (B) (probe 3 in Fig. 2). Lanes 1, RCAD1 (Bph− Etb−); lanes 2, RCD1 (Bph− Etb+); lanes 3, RHA1 (Bph+ Etb+).
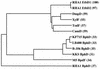
FIG. 9
Phylogenetic tree for EtbD1, EtbD2, BphD, and related hydrolases. The tree was deduced from pairwise alignments of amino acid sequences by using an unweighted pair group method. Enzyme abbreviations: DmpD, dmpD product of Pseudomonas putida CF600 (22); XylF, xylF product of P. putida mt-2 (14); TodF, todF product of P. putida F1 (21); CumD, cumD product of Pseudomonas fluorescens IP01 (8); KF715 BphD, bphD product of P. putida KF715 (12); LB400 BphD, bphD product of Pseudomonas sp. strain LB400 (13); B-356 BphD, bphD product of Comamonas testosteroni B-356 (1); KKS BphD, bphD product of Pseudomonas sp. strain KKS102 (16); M5 BpdF, bpdF product of Rhodococcus sp. strain M5 (18). The percentages of amino acid sequence identity with EtbD1 are shown in parentheses.
Similar articles
- The bphDEF meta-cleavage pathway genes involved in biphenyl/polychlorinated biphenyl degradation are located on a linear plasmid and separated from the initial bphACB genes in Rhodococcus sp. strain RHA1.
Masai E, Sugiyama K, Iwashita N, Shimizu S, Hauschild JE, Hatta T, Kimbara K, Yano K, Fukuda M. Masai E, et al. Gene. 1997 Mar 10;187(1):141-9. doi: 10.1016/s0378-1119(96)00748-2. Gene. 1997. PMID: 9073078 - Structural alteration of linear plasmids encoding the genes for polychlorinated biphenyl degradation in Rhodococcus strain RHA1.
Fukuda M, Shimizu S, Okita N, Seto M, Masai E. Fukuda M, et al. Antonie Van Leeuwenhoek. 1998 Jul-Oct;74(1-3):169-73. doi: 10.1023/a:1001732718159. Antonie Van Leeuwenhoek. 1998. PMID: 10068798 - Characterization of biphenyl catabolic genes of gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1.
Masai E, Yamada A, Healy JM, Hatta T, Kimbara K, Fukuda M, Yano K. Masai E, et al. Appl Environ Microbiol. 1995 Jun;61(6):2079-85. doi: 10.1128/aem.61.6.2079-2085.1995. Appl Environ Microbiol. 1995. PMID: 7793929 Free PMC article. - Cloning and characterization of benzoate catabolic genes in the gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1.
Kitagawa W, Miyauchi K, Masai E, Fukuda M. Kitagawa W, et al. J Bacteriol. 2001 Nov;183(22):6598-606. doi: 10.1128/JB.183.22.6598-6606.2001. J Bacteriol. 2001. PMID: 11673430 Free PMC article. - Crystal structure of 2-hydroxyl-6-oxo-6-phenylhexa-2,4-dienoic acid (HPDA) hydrolase (BphD enzyme) from the Rhodococcus sp. strain RHA1 of the PCB degradation pathway.
Nandhagopal N, Yamada A, Hatta T, Masai E, Fukuda M, Mitsui Y, Senda T. Nandhagopal N, et al. J Mol Biol. 2001 Jun 22;309(5):1139-51. doi: 10.1006/jmbi.2001.4737. J Mol Biol. 2001. PMID: 11399084
Cited by
- Functional characterization of a catabolic plasmid from polychlorinated- biphenyl-degrading Rhodococcus sp. strain RHA1.
Warren R, Hsiao WW, Kudo H, Myhre M, Dosanjh M, Petrescu A, Kobayashi H, Shimizu S, Miyauchi K, Masai E, Yang G, Stott JM, Schein JE, Shin H, Khattra J, Smailus D, Butterfield YS, Siddiqui A, Holt R, Marra MA, Jones SJ, Mohn WW, Brinkman FS, Fukuda M, Davies J, Eltis LD. Warren R, et al. J Bacteriol. 2004 Nov;186(22):7783-95. doi: 10.1128/JB.186.22.7783-7795.2004. J Bacteriol. 2004. PMID: 15516593 Free PMC article. - Multiple-subunit genes of the aromatic-ring-hydroxylating dioxygenase play an active role in biphenyl and polychlorinated biphenyl degradation in Rhodococcus sp. strain RHA1.
Iwasaki T, Miyauchi K, Masai E, Fukuda M. Iwasaki T, et al. Appl Environ Microbiol. 2006 Aug;72(8):5396-402. doi: 10.1128/AEM.00298-06. Appl Environ Microbiol. 2006. PMID: 16885291 Free PMC article. - Novel pathway of salicylate degradation by Streptomyces sp. strain WA46.
Ishiyama D, Vujaklija D, Davies J. Ishiyama D, et al. Appl Environ Microbiol. 2004 Mar;70(3):1297-306. doi: 10.1128/AEM.70.3.1297-1306.2004. Appl Environ Microbiol. 2004. PMID: 15006746 Free PMC article. - Plasmid pCAR3 contains multiple gene sets involved in the conversion of carbazole to anthranilate.
Urata M, Uchimura H, Noguchi H, Sakaguchi T, Takemura T, Eto K, Habe H, Omori T, Yamane H, Nojiri H. Urata M, et al. Appl Environ Microbiol. 2006 May;72(5):3198-205. doi: 10.1128/AEM.72.5.3198-3205.2006. Appl Environ Microbiol. 2006. PMID: 16672458 Free PMC article. - Roles of ring-hydroxylating dioxygenases in styrene and benzene catabolism in Rhodococcus jostii RHA1.
Patrauchan MA, Florizone C, Eapen S, Gómez-Gil L, Sethuraman B, Fukuda M, Davies J, Mohn WW, Eltis LD. Patrauchan MA, et al. J Bacteriol. 2008 Jan;190(1):37-47. doi: 10.1128/JB.01122-07. Epub 2007 Oct 26. J Bacteriol. 2008. PMID: 17965160 Free PMC article.
References
- Ahmad D, Fraser J, Sylvestre M, Larose A, Khan A, Bergeron J, Juteau J M, Sondossi M. Sequence of the bphD gene encoding 2-hydroxy-6-oxo-(phenyl/chlorophenyl)hexa-2,4-dienoic acid (HOP/cPDA) hydrolase involved in the biphenyl/polychlorinated biphenyl degradation pathway in Comamonas testosteroni: evidence suggesting involvement of Ser112 in catalytic activity. Gene. 1995;156:69–74. - PubMed
- Arand M, Grant D F, Beetham J K, Friedberg T, Oesch F, Hammock B D. Sequence similarity of mammalian epoxide hydrolases to the bacterial haloalkane dehalogenase and other related proteins. FEBS Lett. 1994;338:251–256. - PubMed
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1990.
- Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases