Establishment of new genetic traits in a microbial biofilm community - PubMed (original) (raw)
Establishment of new genetic traits in a microbial biofilm community
B B Christensen et al. Appl Environ Microbiol. 1998 Jun.
Abstract
Conjugational transfer of the TOL plasmid (pWWO) was analyzed in a flow chamber biofilm community engaged in benzyl alcohol degradation. The community consisted of three species, Pseudomonas putida RI, Acinetobacter sp. strain C6, and an unidentified isolate, D8. Only P. putida RI could act as a recipient for the TOL plasmid. Cells carrying a chromosomally integrated lacIq gene and a lacp-gfp-tagged version of the TOL plasmid were introduced as donor strains in the biofilm community after its formation. The occurrence of plasmid-carrying cells was analyzed by viable-count-based enumeration of donors and transconjugants. Upon transfer of the plasmids to the recipient cells, expression of green fluorescence was activated as a result of zygotic induction of the gfp gene. This allowed a direct in situ identification of cells receiving the gfp-tagged version of the TOL plasmid. Our data suggest that the frequency of horizontal plasmid transfer was low, and growth (vertical transfer) of the recipient strain was the major cause of plasmid establishment in the biofilm community. Employment of scanning confocal laser microscopy on fixed biofilms, combined with simultaneous identification of P. putida cells and transconjugants by 16S rRNA hybridization and expression of green fluorescence, showed that transconjugants were always associated with noninfected P. putida RI recipient microcolonies. Pure colonies of transconjugants were never observed, indicating that proliferation of transconjugant cells preferentially took place on preexisting P. putida RI microcolonies in the biofilm.
Figures
FIG. 1
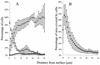
Quantitative analysis showing the spatial distribution of the different organisms of the mixed-culture biofilm sampled at day 7. (A) Vertical profile through the biofilm showing percentage of Acinetobacter sp. strain C6 (▵), P. putida RI (◊), and isolate D8 (□) relative to the total number of cells targeted with the general eubacterial 16S rRNA probe (EUB338 [1]). Each profile is an average of three images (each consisting of 31 optical sections) captured at three random locations in the biofilm. (B) Relative area covered by cells in each section, taken as an average over six images. Standard deviations are indicated by error bars.
FIG. 2
Time course analysis of the distribution of donor cells, P. putida RI, and transconjugants relative to the total number of cells collected from flow channel effluents. The donor (P. putida RI/TOL_gfp_mut3b) was introduced at day 2 in three different densities: 5 · 104 (▴), 5 · 105 (▪), and 5 · 106 (⧫) CFU/ml. Total counts (A) were enumerated on pure LB broth plates. The P. putida RI cells (Nalr) (B), donor cells (Rifr) (C), and transconjugants (Nalr Kmr) (D) were enumerated on LB broth plates containing the appropriate antibiotics, and the numbers were taken relative to the total counts. Each data point is the average of two independent experiments.
FIG. 3
Time course analysis of the distribution of donor cells, P. putida RI, and transconjugants relative to the total number of cells collected from flow channel effluents. The donor (P. putida KT2442/TOL_gfp_mut3b) was introduced at day 2 in three different concentrations: 5 · 106 (▴), 5 · 107 (▪), and 5 · 108 (⧫) CFU/ml. Total counts (A) were enumerated on pure LB broth plates. The P. putida RI cells (Nalr) (B), donor cells (Rifr) (C), and transconjugants (Nalr Kmr) (D) were enumerated on LB broth plates containing the appropriate antibiotics, and the numbers were taken relative to the total counts. Each data point is the average of two independent experiments.
FIG. 4
Time course analysis of the distribution of donor cells, P. putida RI, and transconjugants relative to the total number of cells collected from flow channel effluents. Without changing the inoculation concentration of the two other isolates in the model community, P. putida RI was introduced in three different concentrations of 105 (▴), 107 (▪), and 108 (⧫) CFU/ml. Donor cells (P. putida KT2442/TOL_gfp_mut3b) (5 · 108 CFU/ml) were introduced at day 2. Total counts (A) were enumerated on pure LB broth plates. The Nalr P. putida RI cells (B), Rifr donor cells (C), and Nalr Kmr transconjugants (D) were enumerated on LB broth plates containing the appropriate antibiotics, and the numbers were taken relative to the total counts. Each point is the average of two individual experiments.
FIG. 5
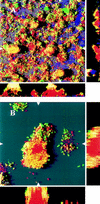
On-line monitoring of transconjugant proliferation on microcolonies in the direction of flow at days 5 and 6 after donor introduction. The white patches are regions with strong green-fluorescent signal (microcolonies with transconjugants), and the gray regions are weak autofluorescent signals emitted from P. putida cells. This signal is easy to distinguish from the strong green-fluorescent signal emitted from cells expressing Gfp and could be used to visualize the location of noninfected microcolonies. On day 5 (A) a strongly green-fluorescent microcolony (solid arrow) was observed and other green-fluorescent microcolonies were located in a region straight downstream from this colony, but not upstream. On day 6 (B) more green-fluorescent colonies were observed. The scale bar also indicates the direction of flow. Open arrows indicate examples of microcolonies which had been infected with transconjugants from day 5 to day 6.
FIG. 6
(A) Spatial distribution of green-fluorescent transconjugants (green or yellow) relative to noninfected P. putida RI cells and Acinetobacter sp. strain C6 in a biofilm analyzed 8 days after introduction of donor cells. The organisms P. putida (red) and Acinetobacter sp. strain C6 (purple) were identified by hybridization. After hybridization, green-fluorescent transconjugants appear as either yellow or green, depending on the ratio between the green Gfp signal and the red hybridization signal. The x-y plot is presented as a SFP, where long shadows indicate a large and/or high microcolony. Shown to the right and below are vertical sections through the biofilm collected at the positions indicated by the white triangles. (B) Magnification of a P. putida colony with green-fluorescent cells covering the surface. Vertical sections through the colony are shown to the right and below. The microcolony is a SFP of a region 10 to 19 μm from the glass surface.
Similar articles
- Bioaugmentation of microbial communities in laboratory and pilot scale sequencing batch biofilm reactors using the TOL plasmid.
Venkata Mohan S, Falkentoft C, Venkata Nancharaiah Y, Sturm BS, Wattiau P, Wilderer PA, Wuertz S, Hausner M. Venkata Mohan S, et al. Bioresour Technol. 2009 Mar;100(5):1746-53. doi: 10.1016/j.biortech.2008.09.048. Epub 2008 Nov 17. Bioresour Technol. 2009. PMID: 19010662 - Plasmid transfer from Pseudomonas putida to the indigenous bacteria on alfalfa sprouts: characterization, direct quantification, and in situ location of transconjugant cells.
Mølbak L, Licht TR, Kvist T, Kroer N, Andersen SR. Mølbak L, et al. Appl Environ Microbiol. 2003 Sep;69(9):5536-42. doi: 10.1128/AEM.69.9.5536-5542.2003. Appl Environ Microbiol. 2003. PMID: 12957943 Free PMC article. - In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members.
Møller S, Sternberg C, Andersen JB, Christensen BB, Ramos JL, Givskov M, Molin S. Møller S, et al. Appl Environ Microbiol. 1998 Feb;64(2):721-32. doi: 10.1128/AEM.64.2.721-732.1998. Appl Environ Microbiol. 1998. PMID: 9464414 Free PMC article. - Dual labeling of Pseudomonas putida with fluorescent proteins for in situ monitoring of conjugal transfer of the TOL plasmid.
Nancharaiah YV, Wattiau P, Wuertz S, Bathe S, Mohan SV, Wilderer PA, Hausner M. Nancharaiah YV, et al. Appl Environ Microbiol. 2003 Aug;69(8):4846-52. doi: 10.1128/AEM.69.8.4846-4852.2003. Appl Environ Microbiol. 2003. PMID: 12902279 Free PMC article.
Cited by
- Green fluorescent protein-labeled monitoring tool to quantify conjugative plasmid transfer between Gram-positive and Gram-negative bacteria.
Arends K, Schiwon K, Sakinc T, Hübner J, Grohmann E. Arends K, et al. Appl Environ Microbiol. 2012 Feb;78(3):895-9. doi: 10.1128/AEM.05578-11. Epub 2011 Dec 2. Appl Environ Microbiol. 2012. PMID: 22138997 Free PMC article. - gfp-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication.
Andersen JB, Heydorn A, Hentzer M, Eberl L, Geisenberger O, Christensen BB, Molin S, Givskov M. Andersen JB, et al. Appl Environ Microbiol. 2001 Feb;67(2):575-85. doi: 10.1128/AEM.67.2.575-585.2001. Appl Environ Microbiol. 2001. PMID: 11157219 Free PMC article. - Fluorescence correlation spectroscopy to study diffusion and reaction of bacteriophages inside biofilms.
Briandet R, Lacroix-Gueu P, Renault M, Lecart S, Meylheuc T, Bidnenko E, Steenkeste K, Bellon-Fontaine MN, Fontaine-Aupart MP. Briandet R, et al. Appl Environ Microbiol. 2008 Apr;74(7):2135-43. doi: 10.1128/AEM.02304-07. Epub 2008 Feb 1. Appl Environ Microbiol. 2008. PMID: 18245240 Free PMC article. - Role of quorum sensing and antimicrobial component production by Serratia plymuthica in formation of biofilms, including mixed biofilms with Escherichia coli.
Moons P, Van Houdt R, Aertsen A, Vanoirbeek K, Engelborghs Y, Michiels CW. Moons P, et al. Appl Environ Microbiol. 2006 Nov;72(11):7294-300. doi: 10.1128/AEM.01708-06. Epub 2006 Sep 22. Appl Environ Microbiol. 2006. PMID: 16997989 Free PMC article. - Single-cell analyses revealed transfer ranges of IncP-1, IncP-7, and IncP-9 plasmids in a soil bacterial community.
Shintani M, Matsui K, Inoue J, Hosoyama A, Ohji S, Yamazoe A, Nojiri H, Kimbara K, Ohkuma M. Shintani M, et al. Appl Environ Microbiol. 2014 Jan;80(1):138-45. doi: 10.1128/AEM.02571-13. Epub 2013 Oct 18. Appl Environ Microbiol. 2014. PMID: 24141122 Free PMC article.
References
- Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459. - PubMed
- Bradley D E, Williams P A. The TOL plasmid is naturally derepressed for transfer. J Gen Microbiol. 1982;128:3019–3024. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases