Purification and characterization of thin pili of IncI1 plasmids ColIb-P9 and R64: formation of PilV-specific cell aggregates by type IV pili - PubMed (original) (raw)
Purification and characterization of thin pili of IncI1 plasmids ColIb-P9 and R64: formation of PilV-specific cell aggregates by type IV pili
T Yoshida et al. J Bacteriol. 1998 Jun.
Abstract
Thin pili of the closely related IncI1 plasmids ColIb-P9 and R64 are required only for liquid mating and belong to the type IV family of pili. They were sedimented by ultracentrifugation from culture medium in which Escherichia coli cells harboring ColIb-P9- or R64-derived plasmids had been grown, and then the pili were purified by CsCl density gradient centrifugation. In negatively stained thin pilus samples, long rods with a diameter of 6 nm, characteristic of type IV pili, were observed under an electron microscope. Gel electrophoretic analysis of purified ColIb-P9 thin pili indicated that thin pili consist of two kinds of proteins, pilin and the PilV protein. Pilin was demonstrated to be the product of the pilS gene. Pilin was first synthesized as a 22-kDa prepilin from the pilS gene and subsequently processed to a 19-kDa protein by the function of the pilU product. The N-terminal amino group of the processed protein was shown to be modified. The C-terminal segments of the pilV products vary among six or seven different types, as a result of shufflon DNA rearrangements of the pilV gene. These PilV proteins were revealed to comprise a minor component of thin pili. Formation of PilV-specific cell aggregates by ColIb-P9 and R64 thin pili was demonstrated and may play an important role in liquid mating.
Figures
FIG. 1
(A) Gene organization of the traA to -D and pilI to -V regions of pKK641-A′ and pCD641-A′. Restriction sites: B, _Bgl_II; P, _Pst_I; V, _Pvu_II; H, _Hin_dIII; Hp, _Hpa_I; C, _Cla_I; and E, _Eco_RI. Below each map, open reading frames are represented by arrows. tra, trc, transfer; pil, formation of thin pilus; shf, shufflon; rci, shufflon-specific recombinase. The solid lines above pKK641-A′ indicate the DNA segments present in pKK691 and pKK692. The crosses on pKK691 mark the locations of the pilS2, pilT2, and pilU2 mutations. (B) Switching of six pilV genes by DNA rearrangement of the ColIb-P9 shufflon. The gene organization of plasmid A′ expressing _pilVA_′ is shown at the top. Black arrows represent the six 19-bp repeats. The three DNA segments are indicated above the diagram. Open reading frames are indicated by stippling. The _rci_-mediated site-specific recombination of plasmid A′ between repeats 2 and 1 caused the inversion of segment A, to yield plasmid A, and the conversion of pilV from _pilVA_′ to pilVA. Subsequent inversion of whole segments resulted in plasmid B′, converting pilV from pilVA to _pilVB_′. Thus, a series of independent or group inversions of the three DNA segments made six pilV genes encoding different C-terminal segments. (C) N-terminal amino acid sequences of the pilS and pilV products. PilS and PilV sequences presented here are identical between ColIb-P9 and R64. The putative cleavage sites of type IV prepilin peptidase are indicated by the arrow. The conserved glycine and glutamic acid in type IV prepilins are indicated by boldface. The N-terminal hydrophobic region is underlined.
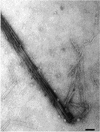
FIG. 2
Electron micrograph of purified ColIb-P9 thin pili. Samples were stained with 4% uranyl acetate. Bar, 50 nm.
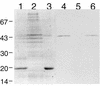
FIG. 3
Protein analysis of ColIb-P9 thin pili. Proteins were separated by SDS-PAGE (17.5% polyacrylamide) and stained with Coomassie brilliant blue (lanes 1 to 3) or analyzed by Western blotting with anti-R64 PilVA′ antiserum (lanes 4 to 6). Lanes 1 and 4, crude thin pilus fraction from E. coli cells harboring pCD641-A′; lanes 2 and 5, crude thin pilus fraction from cells without plasmid; lanes 3 and 6, purified thin pilus fraction. The locations of molecular mass markers (in kilodaltons) are indicated on the left.
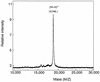
FIG. 4
MALDI/TOFMS spectrum of mature ColIb-P9 pilin. The molecular weight of pilin was estimated with horse heart myoglobin (16,951) and human serum albumin (66,402) as external standards.
FIG. 5
Processing of the R64 pilS product. Whole proteins of E. coli cells harboring pKK691, pKK691 pilS2, pKK691 pilT2, pKK691 pilU2, and pKK692 were separated by SDS-PAGE and subjected to Western blot analysis with antipilin antiserum. Lanes 1 to 5, E. coli cells harboring pKK691, pKK691 pilS2, pKK691 pilT2, pKK691 pilU2, and pKK692, respectively; lane 6, crude thin pilus fraction from cells harboring pKK641-A′; lane 7, cells without plasmid. Numbers on the left are the sizes (in kilodaltons) of marker proteins.
FIG. 6
Detection of the six different PilV proteins in the thin pilus fractions of E. coli cells harboring six pCD641 series plasmids. Crude thin pilus fractions were separated by SDS-PAGE (12.5% polyacrylamide) and subjected to Western blot analysis with anti-PilVA′ antiserum. Lanes 1 to 6, thin pilus fraction from E. coli cells harboring pCD641-A, pCD641-A′, pCD641-B, pCD641-B′, pCD641-C, and pCD641-C′, respectively; lane 7, control from cells without plasmid. Numbers on the left are the sizes (in kilodaltons) of marker proteins.
FIG. 7
Colony and cell morphology of E. coli K-12 and C strains harboring pCD641 series plasmids expressing one of the pilVA, _pilVA_′, pilVB, _pilVB_′, pilVC, and _pilVC_′ genes. pCD641 series plasmids carrying one of these genes were introduced into E. coli strains K-12 and C. Cells were grown on LB agar or in LB media. The cells were photographed under a Nikon microscope. Bars: colonies, 1 cm; cells, 10 μm.
Similar articles
- Mutational analysis of plasmid R64 thin pilus prepilin: the entire prepilin sequence is required for processing by type IV prepilin peptidase.
Horiuchi T, Komano T. Horiuchi T, et al. J Bacteriol. 1998 Sep;180(17):4613-20. doi: 10.1128/JB.180.17.4613-4620.1998. J Bacteriol. 1998. PMID: 9721303 Free PMC article. - The plasmid R64 thin pilus identified as a type IV pilus.
Kim SR, Komano T. Kim SR, et al. J Bacteriol. 1997 Jun;179(11):3594-603. doi: 10.1128/jb.179.11.3594-3603.1997. J Bacteriol. 1997. PMID: 9171405 Free PMC article. - DNA rearrangement of the shufflon determines recipient specificity in liquid mating of IncI1 plasmid R64.
Komano T, Kim SR, Yoshida T, Nisioka T. Komano T, et al. J Mol Biol. 1994 Oct 14;243(1):6-9. doi: 10.1006/jmbi.1994.1625. J Mol Biol. 1994. PMID: 7932741 Review. - Mating variation by DNA inversions of shufflon in plasmid R64.
Komano T, Kim SR, Yoshida T. Komano T, et al. Adv Biophys. 1995;31:181-93. doi: 10.1016/0065-227x(95)99391-2. Adv Biophys. 1995. PMID: 7625273 Review.
Cited by
- Haloferax volcanii flagella are required for motility but are not involved in PibD-dependent surface adhesion.
Tripepi M, Imam S, Pohlschröder M. Tripepi M, et al. J Bacteriol. 2010 Jun;192(12):3093-102. doi: 10.1128/JB.00133-10. Epub 2010 Apr 2. J Bacteriol. 2010. PMID: 20363933 Free PMC article. - How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains.
Brzuszkiewicz E, Brüggemann H, Liesegang H, Emmerth M, Olschläger T, Nagy G, Albermann K, Wagner C, Buchrieser C, Emody L, Gottschalk G, Hacker J, Dobrindt U. Brzuszkiewicz E, et al. Proc Natl Acad Sci U S A. 2006 Aug 22;103(34):12879-84. doi: 10.1073/pnas.0603038103. Epub 2006 Aug 15. Proc Natl Acad Sci U S A. 2006. PMID: 16912116 Free PMC article. - Neisseria gonorrhoeae PilV, a type IV pilus-associated protein essential to human epithelial cell adherence.
Winther-Larsen HC, Hegge FT, Wolfgang M, Hayes SF, van Putten JP, Koomey M. Winther-Larsen HC, et al. Proc Natl Acad Sci U S A. 2001 Dec 18;98(26):15276-81. doi: 10.1073/pnas.261574998. Proc Natl Acad Sci U S A. 2001. PMID: 11752467 Free PMC article. - Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss Army knives.
Berry JL, Pelicic V. Berry JL, et al. FEMS Microbiol Rev. 2015 Jan;39(1):134-54. doi: 10.1093/femsre/fuu001. Epub 2014 Dec 4. FEMS Microbiol Rev. 2015. PMID: 25793961 Free PMC article. Review.
References
- Bradley D E. Derepressed plasmids of incompatibility group I1 determine two different morphological forms of pilus. Plasmid. 1983;9:331–334. - PubMed
- Bradley D E. Characterization and function of thick and thin conjugative pili determined by transfer-derepressed plasmids of incompatibility group I1, I2, I5, B, K, and Z. J Gen Microbiol. 1984;130:1489–1502. - PubMed
- Date T, Inuzuka M, Tomoeda M. Purification and characterization of F pili from Escherichia coli. Biochemistry. 1977;16:5579–5585. - PubMed
- Firth N, Ippen-Ihler K, Skurray R A. Structure and function of the F factor and mechanism of conjugation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 2377–2401.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources