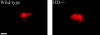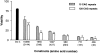The influence of huntingtin protein size on nuclear localization and cellular toxicity - PubMed (original) (raw)
The influence of huntingtin protein size on nuclear localization and cellular toxicity
A S Hackam et al. J Cell Biol. 1998.
Abstract
Huntington disease is an autosomal dominant neurodegenerative disorder caused by the pathological expansion of a polyglutamine tract. In this study we directly assess the influence of protein size on the formation and subcellular localization of huntingtin aggregates. We have created numerous deletion constructs expressing successively smaller fragments of huntingtin and show that these smaller proteins containing 128 glutamines form both intranuclear and perinuclear aggregates. In contrast, larger NH2-terminal fragments of huntingtin proteins with 128 glutamines form exclusively perinuclear aggregates. These aggregates can form in the absence of endogenous huntingtin. Furthermore, expression of mutant huntingtin results in increased susceptibility to apoptotic stress that is greater with decreasing protein length and increasing polyglutamine size. As both intranuclear and perinuclear aggregates are clearly associated with increased cellular toxicity, this supports an important role for toxic polyglutamine-containing fragments forming aggregates and playing a key role in the pathogenesis of Huntington disease.
Figures
Figure 1
(a) Diagrammatic representation of the cDNA expression constructs used in this study. Bars represent huntingtin protein, with amino acids numbered according to GenBank/EMBL/DDBJ accession number L27350. The locations of the polyglutamine tract (Q) are indicated. The names of the constructs, 436, 771, 989, 1597, 1955, and 10366 correspond to the nucleotide position of the translation termination codon. The amino acid length of each protein is also indicated at the right. (b) Western blot of huntingtin truncation constructs. HEK 293T cells were transfected with the 771, 989, 1597, 1955, and 10366 constructs, containing 15 glutamines. 36 h after transfection, cell lysates were prepared. Equal amounts of protein were loaded on a 5–20% SDS-PAGE gradient gel, and Western blotted using anti-huntingtin antibody BKP1, which recognizes the NH2 terminus. Multiple bands in the lanes represent different conformations of the protein or huntingtin multimers. The amino acid length (aa) of each expressed protein is also indicated.
Figure 1
(a) Diagrammatic representation of the cDNA expression constructs used in this study. Bars represent huntingtin protein, with amino acids numbered according to GenBank/EMBL/DDBJ accession number L27350. The locations of the polyglutamine tract (Q) are indicated. The names of the constructs, 436, 771, 989, 1597, 1955, and 10366 correspond to the nucleotide position of the translation termination codon. The amino acid length of each protein is also indicated at the right. (b) Western blot of huntingtin truncation constructs. HEK 293T cells were transfected with the 771, 989, 1597, 1955, and 10366 constructs, containing 15 glutamines. 36 h after transfection, cell lysates were prepared. Equal amounts of protein were loaded on a 5–20% SDS-PAGE gradient gel, and Western blotted using anti-huntingtin antibody BKP1, which recognizes the NH2 terminus. Multiple bands in the lanes represent different conformations of the protein or huntingtin multimers. The amino acid length (aa) of each expressed protein is also indicated.
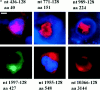
Figure 2
(a) Immunofluorescence of 293T cells transfected with the 436, 771, 989, 1597, 1955, and 10366 constructs, each containing 128 glutamines. 36–40 h after transfection, the cells were processed for immunofluorescence using mAb 2166 (red) or BKP1 (green) to detect huntingtin. The cells were counterstained with the DNA stain DAPI (blue) to label the nucleus (the DAPI stain is not visible in the cells shown for 1597-128 and 10366-128). Huntingtin located in the nucleus appears pink as the red stain is overlaid with blue. Huntingtin from the 436, 771, and 989 constructs (top row) appears as large intranuclear aggregates. Huntingtin from the 1597, 1955, and 10366 constructs (bottom row) show perinuclear aggregates and diffuse cytoplasmic stain. (b) Size-exclusion chromatography of a lysate from 293T cells transfected with HD1955-128 and treated with tamoxifen. 1-ml fractions were collected at 4°C, and samples were immediately analyzed by reducing SDS-PAGE and Western blotting with BKP1 antibody. Large molecular weight complexes were not retained by the column and flowed through into the void volume. Huntingtin aggregates isolated in the void volume were observed as species that did not denature upon SDS-PAGE and did not exit the stacking gel (V o = 42 ml; lanes 2 and 3). Under the same conditions, no aggregation was observed in the whole cell lysate (lane 1) nor in the monomeric huntingtin fractions (V = 72 ml; lanes 4 and 5). Bar in a: (436-128) 11.3 μm; (771-128) 5.31 μm; (989-128) 11.2 μm; (1597-128) 11.97 μm; (1955-128) 12 μm; (10366-128) 11.4 μm.
Figure 2
(a) Immunofluorescence of 293T cells transfected with the 436, 771, 989, 1597, 1955, and 10366 constructs, each containing 128 glutamines. 36–40 h after transfection, the cells were processed for immunofluorescence using mAb 2166 (red) or BKP1 (green) to detect huntingtin. The cells were counterstained with the DNA stain DAPI (blue) to label the nucleus (the DAPI stain is not visible in the cells shown for 1597-128 and 10366-128). Huntingtin located in the nucleus appears pink as the red stain is overlaid with blue. Huntingtin from the 436, 771, and 989 constructs (top row) appears as large intranuclear aggregates. Huntingtin from the 1597, 1955, and 10366 constructs (bottom row) show perinuclear aggregates and diffuse cytoplasmic stain. (b) Size-exclusion chromatography of a lysate from 293T cells transfected with HD1955-128 and treated with tamoxifen. 1-ml fractions were collected at 4°C, and samples were immediately analyzed by reducing SDS-PAGE and Western blotting with BKP1 antibody. Large molecular weight complexes were not retained by the column and flowed through into the void volume. Huntingtin aggregates isolated in the void volume were observed as species that did not denature upon SDS-PAGE and did not exit the stacking gel (V o = 42 ml; lanes 2 and 3). Under the same conditions, no aggregation was observed in the whole cell lysate (lane 1) nor in the monomeric huntingtin fractions (V = 72 ml; lanes 4 and 5). Bar in a: (436-128) 11.3 μm; (771-128) 5.31 μm; (989-128) 11.2 μm; (1597-128) 11.97 μm; (1955-128) 12 μm; (10366-128) 11.4 μm.
Figure 3
Immunofluorescence of 293T cell transfected with the COOH-terminal huntingtin construct (amino acids 585–3,144). Transfected cells were treated with tamoxifen 36–40 h after transfection and processed for immunofluorescence using mAb 2172 to detect huntingtin (red). The protein was localized exclusively throughout the cytoplasm. Bar, 4.42 μm.
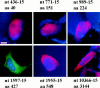
Figure 4
Immunofluorescence of 293T cells transfected with the 436, 771, 989, 1597, 1955, and 10366 constructs, each containing 15 glutamines. The transfected cells were treated with tamoxifen 36-40 h after transfection and mAb 2166 (red) or BKP1 (green) was used to detect huntingtin. The cells were counterstained with the DNA stain DAPI (blue) (the DAPI stain is not visible in the 1597-15 panel). Huntingtin containing 15 repeats does not form aggregates but can be localized in the nucleus depending on the protein size. Huntingtin is localized entirely in the nucleus in the 436 and 771 constructs in the cells shown. Bar: (436-15) 8.87 μm; (771-15) 11.98 μm; (989-15) 16.18 μm; (1597-15) 16.04 μm; (1955-15) 15.2 μm; (10366-15) 21.37 μm.
Figure 5
Immunofluorescence of wild-type and null (HD −/−) ES cells transfected with 1955-128. The cells were treated with tamoxifen and immunostained with mAb 2166. Perinuclear aggregates were evident in both cell lines, indicating that the presence of endogenous huntingtin is not required for the formation of aggregates by transfected 1955-128. Bar: (Wild-type) 2.82 μm; (HD −/−) 2.87 μm.
Figure 6
Increasing toxicity with decreasing huntingtin length. In the presence of tamoxifen, progressive truncation of huntingtin resulted in increased cellular toxicity (P < 0.01). Interestingly, this trend was seen for proteins containing both 15 and 128 polyglutamines (P < 0.01). 293T cells were transfected with each huntingtin construct, or LacZ control, and cell viability in response to apoptotic stress was measured using a modified MTT assay and presented relative to control (untransfected). All wells were transfected with 0.1 μg DNA except for those transfected with the 436-15 and 128 constructs, where 0.08 μg of DNA was used because all the cells died with 0.1 μg DNA. Results for the 436 construct are normalized, taking into account decreased DNA concentration. Asterisks indicate the level of significance between huntingtin containing 15 and 128 glutamines: * P < 0.01; ** P < 0.001.
Similar articles
- Length of huntingtin and its polyglutamine tract influences localization and frequency of intracellular aggregates.
Martindale D, Hackam A, Wieczorek A, Ellerby L, Wellington C, McCutcheon K, Singaraja R, Kazemi-Esfarjani P, Devon R, Kim SU, Bredesen DE, Tufaro F, Hayden MR. Martindale D, et al. Nat Genet. 1998 Feb;18(2):150-4. doi: 10.1038/ng0298-150. Nat Genet. 1998. PMID: 9462744 - Truncated N-terminal fragments of huntingtin with expanded glutamine repeats form nuclear and cytoplasmic aggregates in cell culture.
Cooper JK, Schilling G, Peters MF, Herring WJ, Sharp AH, Kaminsky Z, Masone J, Khan FA, Delanoy M, Borchelt DR, Dawson VL, Dawson TM, Ross CA. Cooper JK, et al. Hum Mol Genet. 1998 May;7(5):783-90. doi: 10.1093/hmg/7.5.783. Hum Mol Genet. 1998. PMID: 9536081 - In vitro evidence for both the nucleus and cytoplasm as subcellular sites of pathogenesis in Huntington's disease.
Hackam AS, Singaraja R, Zhang T, Gan L, Hayden MR. Hackam AS, et al. Hum Mol Genet. 1999 Jan;8(1):25-33. doi: 10.1093/hmg/8.1.25. Hum Mol Genet. 1999. PMID: 9887328 - Evidence for both the nucleus and cytoplasm as subcellular sites of pathogenesis in Huntington's disease in cell culture and in transgenic mice expressing mutant huntingtin.
Hackam AS, Hodgson JG, Singaraja R, Zhang T, Gan L, Gutekunst CA, Hersch SM, Hayden MR. Hackam AS, et al. Philos Trans R Soc Lond B Biol Sci. 1999 Jun 29;354(1386):1047-55. doi: 10.1098/rstb.1999.0457. Philos Trans R Soc Lond B Biol Sci. 1999. PMID: 10434304 Free PMC article. Review. - Huntingtin processing in pathogenesis of Huntington disease.
Qin ZH, Gu ZL. Qin ZH, et al. Acta Pharmacol Sin. 2004 Oct;25(10):1243-9. Acta Pharmacol Sin. 2004. PMID: 15456523 Review.
Cited by
- Emerging Therapies for Huntington's Disease - Focus on N-Terminal Huntingtin and Huntingtin Exon 1.
van der Bent ML, Evers MM, Vallès A. van der Bent ML, et al. Biologics. 2022 Sep 30;16:141-160. doi: 10.2147/BTT.S270657. eCollection 2022. Biologics. 2022. PMID: 36213816 Free PMC article. Review. - A survey of protein interactions and posttranslational modifications that influence the polyglutamine diseases.
Johnson SL, Tsou WL, Prifti MV, Harris AL, Todi SV. Johnson SL, et al. Front Mol Neurosci. 2022 Sep 14;15:974167. doi: 10.3389/fnmol.2022.974167. eCollection 2022. Front Mol Neurosci. 2022. PMID: 36187346 Free PMC article. Review. - Ataxin 1, a SCA1 neurodegenerative disorder protein, is functionally linked to the silencing mediator of retinoid and thyroid hormone receptors.
Tsai CC, Kao HY, Mitzutani A, Banayo E, Rajan H, McKeown M, Evans RM. Tsai CC, et al. Proc Natl Acad Sci U S A. 2004 Mar 23;101(12):4047-52. doi: 10.1073/pnas.0400615101. Epub 2004 Mar 11. Proc Natl Acad Sci U S A. 2004. PMID: 15016912 Free PMC article. - Genetics and neuropathology of Huntington's disease.
Reiner A, Dragatsis I, Dietrich P. Reiner A, et al. Int Rev Neurobiol. 2011;98:325-72. doi: 10.1016/B978-0-12-381328-2.00014-6. Int Rev Neurobiol. 2011. PMID: 21907094 Free PMC article. Review. - From pathways to targets: understanding the mechanisms behind polyglutamine disease.
Weber JJ, Sowa AS, Binder T, Hübener J. Weber JJ, et al. Biomed Res Int. 2014;2014:701758. doi: 10.1155/2014/701758. Epub 2014 Sep 21. Biomed Res Int. 2014. PMID: 25309920 Free PMC article. Review.
References
- Andrew SE, Goldberg YP, Hayden MR. Commentary: rethinking genotype and phenotype correlations in polyglutamine expansion disorders. Hum Mol Genet. 1997;6:2005–2010. - PubMed
- Becher, M.W., J.A. Kotzuk, A.H. Sharp, S.W. Davies, G.P. Bates, D.L. Price, and C.A. Ross. 1997. Intranuclear neuronal inclusions in Huntington's disease and dentatorubral and palidoluysian atrophy: correlation between the density of inclusions and IT15 CAG triplet repeat length. Neurobiol. Dis. In press. - PubMed
- Carmichael J, DeGraff WG, Gazdar AF, Minna JD, Mitchell JB. Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of chemosensitivity testing. Cancer Res. 1987;47:936–942. - PubMed
- Chen CF, Li S, Chen Y, Chen PL, Sharp ZD, Lee WH. The nuclear localization sequences of the BRCA1 protein interact with the importin-α subunit of the nuclear transport signal receptor. J Biol Chem. 1996;271:32863–32868. - PubMed
- Cooper JK, Schilling G, Peters MF, Herring WJ, Sharp AH, Kaminsky Z, Masone J, Khan FA, Delanoy M, Borchelt DR, et al. Truncated N-terminal fragments of huntingtin with expanded glutamine repeats form nuclear and cytoplasmic aggregates in cell culture. Hum Mol Genet. 1998;7:783–790. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials