Assembly, subunit composition, and footprint of human DNA repair excision nuclease - PubMed (original) (raw)
Assembly, subunit composition, and footprint of human DNA repair excision nuclease
M Wakasugi et al. Proc Natl Acad Sci U S A. 1998.
Abstract
The assembly and composition of human excision nuclease were investigated by electrophoretic mobility shift assay and DNase I footprinting. Individual repair factors or any combination of up to four repair factors failed to form DNA-protein complexes of high specificity and stability. A stable complex of high specificity can be detected only when XPA/RPA, transcription factor IIH, XPC.HHR23B, and XPG and ATP are present in the reaction mixture. The XPF.ERCC1 heterodimer changes the electrophoretic mobility of the DNA-protein complex formed with the other five repair factors, but it does not confer additional specificity. By using proteins with peptide tags or antibodies to the repair factors in electrophoretic mobility shift assays, it was found that XPA, replication protein A, transcription factor IIH, XPG, and XPF.excision repair cross-complementing 1 but not XPC.HHR23B were present in the penultimate and ultimate dual incision complexes. Thus, it appears that XPC.HHR23B is a molecular matchmaker that participates in the assembly of the excision nuclease but is not present in the ultimate dual incision complex. The excision nuclease makes an assymmetric DNase I footprint of approximately 30 bp around the damage and increases the DNase I sensitivity of the DNA on both sides of the footprint.
Figures
Figure 1
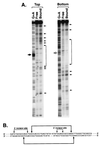
Damage-specific binding and excision. (A) Binding of human excision repair factors to damaged DNA. Repair factors were incubated for 30 min with either a 136-bp control DNA or a duplex containing (–4) photoproduct and were analyzed on a nondenaturing gel (lanes 1–14). (B) Excision reaction by repair factors used in binding assay. The (–4) substrate was incubated for 2 h with the five repair factors (RFI–V: XPA, RPA, TFIIH, XPC, XPG) at the concentration used in binding experiment and XPF⋅ERCC1 (RFVI). Excision products were analyzed on 8% denaturing polyacrylamide gel. Quantitative analysis shows that 93% of the damage was excised.
Figure 2
Detection of preincision complexes 1 and 2 by electrophoretic mobility shift assay. (A) Weak PIC1 forms with RFI-IV (XPA, RPA, TFIIH, and XPC) (lane 2), which is stabilized by XPG to form PIC2 (lane 3). No PIC2 can be detected with unmodified DNA (lane 5). (B) Formation of PIC2 requires all five repair factors. (C) PIC2 formation is ATP-dependent. Reaction mixtures were incubated in the presence or absence of ATP and separated on a 3.5% nondenaturing polyacrylamide gel as indicated. Open arrow shows the binding by a minor contaminant in the TFIIH factor.
Figure 3
Detection of preincision complex 3 by electrophoretic mobility shift assay. The substrate was incubated with RFI-IV (XPA, RPA, TFIIH, XPC) and either wild-type or mutant XPG at 30°C to form PIC2 (1st incubation), then XPF⋅ERCC1 was added on ice as indicated (second incubation), and the DNA–protein complexes were analyzed by electrophoretic mobility shift assay.
Figure 4
Detection of repair factors in PIC3. (A) The PIC3 formed with MBP-XPA migrates slower than the complex formed with smaller (His)6–XPA. (B) Antibodies to p70 and p34 subunits of RPA supershift PIC3. (C) Polyclonal antibodies to the XPB subunit of TFIIH supershift (lane 4); monoclonal XPB and polyclonal p62 antibodies disrupt PIC3 (lanes 3 and 5). PIC3* indicates supershifted PIC3.
Figure 5
Evidence for lack of XPC in PIC3. (A) PIC3s formed with either XPC⋅HHR23B or XPC have the same electrophoretic mobility. (B) Anti-XPC antibodies do not supershift PIC3. (C and D) Band mobility shift assays with T<>T(5′-10) “bubble” substrate. (C) PIC3s formed with (lane 2) and without (lane 3) XPC⋅HHR23B have the same electrophoretic mobility. (D) XPC⋅HHR23B (lane 3) and XPG (lane 4) cause supershift in PIC1 formed without XPC⋅HHR23B, but the combination of the two does not cause further retardation (lane 5).
Figure 6
DNase I footprint of PIC2. The 136-bp substrate with 5′ label either in the strand containing the (–4) photoproduct (top strand) or in the complementary strand (bottom strand) was incubated with the five repair factors (no XPF⋅ERCC1) and digested with DNase I, and then bound and unbound fractions were separated on nondenaturing polyacrylamide gels; DNA was eluted from the free and bound fractions and analyzed on 6% denaturing polyacrylamide gels. (A) Footprinting gels along with G+A sequence ladder. Solid and open arrows show the site of (–4) photoproduct. Brackets and arrow indicate protected region and prominent hypersensitive sites. (B) Schematic illustration of footprint of human excinuclease indicating incision sites and protected region.
Figure 7
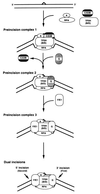
Model for reaction mechanism of human excinuclease. XPC⋅HHR23B is a molecular matchmaker that is not present in either the penultmate or the ultimate dual incision complex. A, B, and D, XPA, XPB, and XPD, respectively; C/23B, XPC⋅HHR23B, G, XPG; and F/E1, XPF⋅ERCC1 complex.
Similar articles
- Mechanism of open complex and dual incision formation by human nucleotide excision repair factors.
Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD. Evans E, et al. EMBO J. 1997 Nov 3;16(21):6559-73. doi: 10.1093/emboj/16.21.6559. EMBO J. 1997. PMID: 9351836 Free PMC article. - Order of assembly of human DNA repair excision nuclease.
Wakasugi M, Sancar A. Wakasugi M, et al. J Biol Chem. 1999 Jun 25;274(26):18759-68. doi: 10.1074/jbc.274.26.18759. J Biol Chem. 1999. PMID: 10373492 - Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome.
Araújo SJ, Nigg EA, Wood RD. Araújo SJ, et al. Mol Cell Biol. 2001 Apr;21(7):2281-91. doi: 10.1128/MCB.21.7.2281-2291.2001. Mol Cell Biol. 2001. PMID: 11259578 Free PMC article. - DNA damage recognition during nucleotide excision repair in mammalian cells.
Wood RD. Wood RD. Biochimie. 1999 Jan-Feb;81(1-2):39-44. doi: 10.1016/s0300-9084(99)80036-4. Biochimie. 1999. PMID: 10214908 Review. - Xeroderma pigmentosum and molecular cloning of DNA repair genes.
Boulikas T. Boulikas T. Anticancer Res. 1996 Mar-Apr;16(2):693-708. Anticancer Res. 1996. PMID: 8687116 Review.
Cited by
- Nucleotide excision repair: a versatile and smart toolkit.
Zhang X, Yin M, Hu J. Zhang X, et al. Acta Biochim Biophys Sin (Shanghai). 2022 May 25;54(6):807-819. doi: 10.3724/abbs.2022054. Acta Biochim Biophys Sin (Shanghai). 2022. PMID: 35975604 Free PMC article. Review. - An alternative form of replication protein a expressed in normal human tissues supports DNA repair.
Kemp MG, Mason AC, Carreira A, Reardon JT, Haring SJ, Borgstahl GE, Kowalczykowski SC, Sancar A, Wold MS. Kemp MG, et al. J Biol Chem. 2010 Feb 12;285(7):4788-97. doi: 10.1074/jbc.M109.079418. Epub 2009 Dec 7. J Biol Chem. 2010. PMID: 19996105 Free PMC article. - Platelet-activating factor receptor agonists mediate xeroderma pigmentosum A photosensitivity.
Yao Y, Harrison KA, Al-Hassani M, Murphy RC, Rezania S, Konger RL, Travers JB. Yao Y, et al. J Biol Chem. 2012 Mar 16;287(12):9311-21. doi: 10.1074/jbc.M111.332395. Epub 2012 Feb 1. J Biol Chem. 2012. PMID: 22303003 Free PMC article. - Molecular architecture and functional dynamics of the pre-incision complex in nucleotide excision repair.
Yu J, Yan C, Paul T, Brewer L, Tsutakawa SE, Tsai CL, Hamdan SM, Tainer JA, Ivanov I. Yu J, et al. Nat Commun. 2024 Oct 1;15(1):8511. doi: 10.1038/s41467-024-52860-y. Nat Commun. 2024. PMID: 39353945 Free PMC article. - The ubiquitin-proteasome system in cancer, a major player in DNA repair. Part 1: post-translational regulation.
Vlachostergios PJ, Patrikidou A, Daliani DD, Papandreou CN. Vlachostergios PJ, et al. J Cell Mol Med. 2009 Sep;13(9B):3006-18. doi: 10.1111/j.1582-4934.2009.00824.x. Epub 2009 Jun 11. J Cell Mol Med. 2009. PMID: 19522845 Free PMC article. Review.
References
- Sancar A. Annu Rev Biochem. 1996;65:43–81. - PubMed
- Mu D, Park C H, Matsunaga T, Hsu D S, Reardon J T, Sancar A. J Biol Chem. 1995;270:2415–2418. - PubMed
- Mu D, Hsu D S, Sancar A. J Biol Chem. 1996;271:8285–8294. - PubMed
- Moggs J G, Yarema K J, Essigmann J M, Wood R D. J Biol Chem. 1996;271:7177–7186. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources