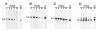Nop5p is a small nucleolar ribonucleoprotein component required for pre-18 S rRNA processing in yeast - PubMed (original) (raw)
Nop5p is a small nucleolar ribonucleoprotein component required for pre-18 S rRNA processing in yeast
P Wu et al. J Biol Chem. 1998.
Abstract
We have identified a novel nucleolar protein, Nop5p, that is essential for growth in Saccharomyces cerevisiae. Monoclonal antibodies B47 and 37C12 recognize Nop5p, which has a predicted size of 57 kDa and possesses a KKX repeat motif at its carboxyl terminus. Truncations that removed the KKX motif were functional and localized to the nucleolus, but conferred slow growth at 37 degreesC. Nop5p shows significant sequence homology with yeast Sik1p/Nop56p, and putative homologues in archaebacteria, plants, and human. Depletion of Nop5p in a GAL-NOP5 strain lengthened the doubling time about 5-fold, and selectively reduced steady-state levels of 40 S ribosomal subunits and 18 S rRNA relative to levels of free 60 S subunits and 25 S rRNA. Northern blotting and primer extension analyses showed that Nop5p depletion impairs processing of 35 S pre-rRNA at the A0 and A2 cleavage sites. Nop5p is associated with the small nucleolar RNAs U3, snR13, U14, and U18. Depletion of Nop5p caused the nucleolar protein Nop1p (yeast fibrillarin) to be localized to the nucleus and cytosol. Also, 37C12 co-immunoprecipitated Nop1p. These results suggest that Nop5p functions with Nop1p in the execution of early pre-rRNA processing steps that lead to formation of 18 S rRNA.
Figures
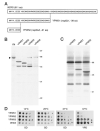
Fig. 1. Immunofluorescence localization
Yeast cells were stained with monoclonal antibodies 37C12 (A) or B47 (E), which were detected with secondary antibody conjugated to Cy3. Cells were also incubated with affinity purified polyclonal antibody (APpAb3) directed against Nop2p and detected with Cy2 conjugated secondary antibody (B and F). DAPI staining and phase-contrast images are also shown (C, D, G, and H). Arrows, crescent-shaped nucleoli. Bars, 10 _μ_m.
Fig. 2. Immunoprecipitation with mAb 37C12
Yeast were labeled with [35S]methionine and a nuclear fraction was prepared for immunoprecipitation. Immunoprecipitates were analyzed by SDS-PAGE. Control immunoprecipitations were done without mAb. 37C12 immunoprecipitates were denatured in SDS, alkylated, renatured, and re-immunoprecipitated with mAb A66 (specific for Nop1p) or no mAb. Supernatants (SN) from the re-immunoprecipitations were trichloroacetic acid-precipitated prior to SDS-PAGE. Molecular weights in kDa.
Fig. 3. Nop5p features
A, an immunoblot of protein extracts from a 37C12 positive λ lysogen (PL) or a control lysogen (CL) induced with isopropyl-1-thio-_β_-
d
-galactopyranoside (+) or not induced (−) was probed with mAb 37C12. The apparent size of the inducible immunoreactive protein (→) is ~140 kDa. A protein of ~67 kDa (*) reacted nonspecifically. B, predicted sequence of Nop5p.
Fig. 4. Localization of HA epitope-tagged Nop5p
A, immunoblot of whole cell protein extracts from strain YPW38 grown in the presence of glucose (lane 1) or galactose (lane 2) probed with mAb 12CA5. A control extract was prepared from YSB25 grown on galactose (lane 3). Molecular weights are in kDa. B, immunofluorescence localization of epitope-tagged Nop5p with mAb 12CA5. Localization of Nop2p with an affinity purified polyclonal antibody (APpAb3), staining of chromatin with DAPI, and a phase-contrast image of the same field of cells is shown. Arrows, crescent-shaped nucleoli. Bar, 10 _μ_m.
Fig. 5. Deletion of the COOH-terminal KKX repeat motif from Nop5p
A, diagram of two COOH-terminal nop5 truncations. The number of deleted amino acids is in parentheses. B, Western blotting of isolated nuclei from BJ2168 (YN) or of nuclear fractions from the strains indicated (YPW45 contains NOP5, YPW46 expresses an HA epitope tagged Nop5p). C, immunoprecipitations with mAb 37C12 from the strains indicated. D, comparison of growth at the indicated temperatures on either minimal glucose (SD) or rich (YPD) media. Serial 10-fold dilutions were replica plated and grown for 10 days at 14 °C, 5 days at 25 °C, or 2 days at 37 °C.
Fig. 6. Alignment of Nop5p with related proteins
Four proteins possess significant sequence similarities with Nop5p: Sik1p/Nop56p from S. cerevisiae (40), and three predicted proteins from C. elegans, A. thaliana, and M. jannaschii (GenPept protein accession numbers 1272634, 2191188, and 2128446, respectively). Searches of the TIGR human sequence data base uncovered 1 tentative human consensus (THC) and 3 expressed sequence tag (EST) sequences similar to Nop5p: THC198801, AA148805, and U56656. These sequences were compiled into a tentative partial sequence that we refer to as a putative human homologue of Nop5p (hNop5p). Alignment was generated by CLUSTAL. Residues identical to the consensus are boxed. Protein sequences that are also similar to Nop5p, but exhibit greater similarity to Sik1p/Nop56p, have been omitted from this alignment.
Fig. 7. NOP5 is an essential gene
A, map of a portion of chromosome XV illustrating the replacement of NOP5 with TRP1. The positions of primers 1–5 are indicated. B, Southern blot of genomic DNA from YSB25, YPW42, or YPW43 digested with _Cla_I (lanes 1–3) or _Xba_I (lanes 5 and 6). Sizes in kb. C, replica platings on media with and without 5-FOA. Serial dilutions (10-fold) were grown at 30 °C for 3 (SD) or 5 (SGal) days. D, growth after shift to YPD or YPGal media. Cultures were diluted to maintain OD600 below 0.5.
Fig. 8. Nop5p is required for 40 S subunit synthesis
W303–1a or YPW48 were grown in YPGal (Gal) or in YPD (Glu) for 4, 8, or 12 h. Cell extracts were separated on linear 10–50% sucrose gradients and analyzed by absorbance measurement at 254 nm (shown in arbitrary units). The positions of the 40 S, 60 S, and 80 S peaks are indicated (↓). Peaks corresponding to polysomes are distributed between the 80 S peak and the bottom of the gradient.
Fig. 9. The major pre-rRNA processing pathway that yields 18 S rRNA in S. cerevisiae
The 35 S primary transcript contains 18 S, 5.8 S, and 25 S rRNA sequences separated by internal transcribed spacers (ITS1 and ITS2) and flanked by externally transcribed spacers (5′-ETS and 3′-ETS). The processing of the 35 S precursor to mature rRNAs involves endonucleolytic and exonucleolytic steps at the specific sites indicated. Cleavage D occurs in the cytoplasm. The relative positions of oligonucleotides 9–13 are indicated. The pathways leading to 5.8 S and 25 S rRNAs are only briefly summarized here (for a recent review, see Ref. 3).
Fig. 10. Nop5p depletion leads to reduced synthesis of 18 S rRNA
A, YPW48 and W303–1a were grown in galactose-containing medium (SG), or in glucose-containing medium (SD) for 24 h. Total cellular RNAs were separated on a 1% glyoxal-agarose gel and stained with ethidium bromide. B, pulse-chase labeling with [_methyl_-3H]methionine. YPW48 was grown in SG medium or SD medium (for 4, 8, or 12 h), labeled for 2 min, and chased for 0, 2, 4, 8, or 12 min. RNAs were separated as in panel A and detected by fluorography. Samples from 8 and 12 h in glucose are overloaded to show the relative amount of 18 S rRNA. C, pulse-chase labeling with [3H]uracil. Chase times of 0, 2, 8, 16, and 32 min were used because of the dynamics of labeling the intracellular uracil pool.
Fig. 11. Northern blot analysis of rRNA processing during Nop5p depletion
YPW48 was cultured in glucose-containing medium for 0, 2, 4, 8, or 12 h. RNAs from 1.5 OD600 units of cells were loaded per lane, separated on a 1% glyoxal-agarose gel, transferred to a positively charged nylon membrane, and hybridized with oligonucleotide probes complementary to ITS1 (oligo 9, panel A), ITS2 (oligo 10, panel B), 5′-ETS (oligo 11, panel C), or the 18 S and 25 S rRNAs (oligos 12 and 13, panel D). The positions of oligonucleotide probes are illustrated in Fig. 9. The minor pathway intermediate 23 S pre-rRNA is indicated (*).
Fig. 12. Primer extension analysis of rRNA processing during Nop5p depletion
YPW48 was shifted to glucose-containing medium and grown for 0, 2, 4, 12, or 24 h. W303–1a (W) was grown for 24 h in the same medium. Primer extension was done using a method employing unlabeled primers (see “Experimental Procedures”). Bands corresponding to processing sites A0, A1, and A2, and the 5′-end of the 35 S pre-rRNA transcript are indicated. The positions of processing sites and the 5′-end of 35 S were determined using DNA sequencing ladders (not shown). Fig. 9 illustrates the positions of primers used (, , and 15).
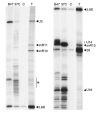
Fig. 13. Immunoprecipitation of small nucleolar RNAs
Isolated nuclei from BJ2168 were solubilized and immunoprecipitations were done with mAb B47 (B47) or mAb 37C12 (37C) or no mAb (C). Immunoprecipitates were extracted to remove proteins, and RNAs were 3′-end labeled, and separated on a denaturing 6% polyacrylamide gel. The top of the gel is shown in the left panel, and the bottom is shown in the right panel. A DNA sequencing ladder was electrophoresed in parallel (not shown) and used to identify the snoRNAs based on their size. RNA bands migrating faster than U18 are predominantly tRNAs.
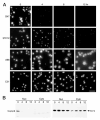
Fig. 14. Nop5p depletion affects the localization of the nucleolar protein Nop1p
A, YPW48 was shifted to glucose-containing medium and grown for 0, 4, 8, or 12 h, after which cells were collected and analyzed by indirect immunofluorescence with the mAbs B47, 37C12, A66 (anti-Nop1p), or C21 (anti-Nsr1p). A secondary antibody Cy3 conjugate was used. Bar, 10 _μ_m. B, YPW48 was grown as described, cells were harvested at the same time points, and crude nuclear and cytoplasmic fractions were prepared and analyzed by Western blotting. The mAbs B47 and D77 (anti-Nop1p) were incubated with the blot simultaneously.
Similar articles
- Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast.
Dunbar DA, Wormsley S, Agentis TM, Baserga SJ. Dunbar DA, et al. Mol Cell Biol. 1997 Oct;17(10):5803-12. doi: 10.1128/MCB.17.10.5803. Mol Cell Biol. 1997. PMID: 9315638 Free PMC article. - Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre-rRNA processing and modification.
Bergès T, Petfalski E, Tollervey D, Hurt EC. Bergès T, et al. EMBO J. 1994 Jul 1;13(13):3136-48. doi: 10.1002/j.1460-2075.1994.tb06612.x. EMBO J. 1994. PMID: 8039506 Free PMC article. - Synthesis and assembly of the box C+D small nucleolar RNPs.
Lafontaine DL, Tollervey D. Lafontaine DL, et al. Mol Cell Biol. 2000 Apr;20(8):2650-9. doi: 10.1128/MCB.20.8.2650-2659.2000. Mol Cell Biol. 2000. PMID: 10733567 Free PMC article. - Mpp10p, a new protein component of the U3 snoRNP required for processing of 18S rRNA precursors.
Baserga SJ, Agentis TM, Wormsley S, Dunbar DA, Lee S. Baserga SJ, et al. Nucleic Acids Symp Ser. 1997;(36):64-7. Nucleic Acids Symp Ser. 1997. PMID: 9478208 Review. - The role of small nucleolar ribonucleoproteins in ribosome synthesis.
Tollervey D, Hurt EC. Tollervey D, et al. Mol Biol Rep. 1990;14(2-3):103-6. doi: 10.1007/BF00360433. Mol Biol Rep. 1990. PMID: 2141891 Review. No abstract available.
Cited by
- Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae.
Kressler D, Linder P, de La Cruz J. Kressler D, et al. Mol Cell Biol. 1999 Dec;19(12):7897-912. doi: 10.1128/MCB.19.12.7897. Mol Cell Biol. 1999. PMID: 10567516 Free PMC article. Review. No abstract available. - The 2'-O-methyltransferase responsible for modification of yeast tRNA at position 4.
Wilkinson ML, Crary SM, Jackman JE, Grayhack EJ, Phizicky EM. Wilkinson ML, et al. RNA. 2007 Mar;13(3):404-13. doi: 10.1261/rna.399607. Epub 2007 Jan 22. RNA. 2007. PMID: 17242307 Free PMC article. - Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli.
Narayanan A, Speckmann W, Terns R, Terns MP. Narayanan A, et al. Mol Biol Cell. 1999 Jul;10(7):2131-47. doi: 10.1091/mbc.10.7.2131. Mol Biol Cell. 1999. PMID: 10397754 Free PMC article. - Protein-protein and protein-RNA contacts both contribute to the 15.5K-mediated assembly of the U4/U6 snRNP and the box C/D snoRNPs.
Schultz A, Nottrott S, Watkins NJ, Lührmann R. Schultz A, et al. Mol Cell Biol. 2006 Jul;26(13):5146-54. doi: 10.1128/MCB.02374-05. Mol Cell Biol. 2006. PMID: 16782898 Free PMC article. - Position within the host intron is critical for efficient processing of box C/D snoRNAs in mammalian cells.
Hirose T, Steitz JA. Hirose T, et al. Proc Natl Acad Sci U S A. 2001 Nov 6;98(23):12914-9. doi: 10.1073/pnas.231490998. Epub 2001 Oct 23. Proc Natl Acad Sci U S A. 2001. PMID: 11606788 Free PMC article.
References
- Tollervey D. Exp. Cell Res. 1996;229:226–232. - PubMed
- Maxwell ES, Fournier MJ. Annu. Rev. Biochem. 1995;64:897–934. - PubMed
- Venema J, Tollervey D. Yeast. 1995;11:1629–1650. - PubMed
- Abou Elela S, Igel H, Ares M. Cell. 1996;85:115–124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases