Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation - PubMed (original) (raw)
Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation
M Onishi et al. Mol Cell Biol. 1998 Jul.
Abstract
STAT (signal transducers and activators of transcription) proteins are transcription factors which are activated by phosphorylation on tyrosine residues upon stimulation by cytokines. Seven members of the STAT family are known, including the closely related STAT5A and STAT5B, which are activated by various cytokines. Except for prolactin-dependent beta-casein production in mammary gland cells, the biological consequences of STAT5 activation in various systems are not clear. We applied PCR-driven random mutagenesis and a retrovirus-mediated expression screening system to identify constitutively active forms of STAT5. By this strategy, we have identified a constitutively active STAT5 mutant which has two amino acid substitutions; one is located upstream of the putative DNA binding domain (H299R), and the other is located in the transactivation domain (S711F). The mutant STAT5 was constitutively phosphorylated on tyrosine residues, localized in the nucleus, and was transcriptionally active. Expression of the mutant STAT5 partially dispenses with interleukin 3 (IL-3) as a growth stimulant of IL-3-dependent cell lines. Further analyses of the mutant STAT5 have demonstrated that both of the mutations are required for nuclear localization, efficient transcriptional activation, and induction of IL-3-independent growth of an IL-3-dependent cell line, Ba/F3, and have indicated that a molecular basis for the constitutive activation is the stability of the phosphorylated form of the mutant STAT5.
Figures
FIG. 1
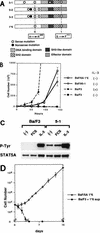
Isolation and characterization of a constitutively active STAT5A. (A) Positions of point mutations of the mutant STAT5A. A constitutively active form of STAT5A1*6 was constructed from subclones 5-1, 5-2, and 5-3, which had been derived from FI Ba/F3 clone 5. Among these subclones, only 5-1 induced IL-3-independent growth of Ba/F3 cells. (B) Proliferation of Ba/F3 cells expressing subclone 5-1 or STAT5A1*6 in the absence of IL-3. BaF/5-1 and BaF/5A1*6 transfectants, which had been selected in the presence of puromycin and mIL-3, were cultured in the absence of mIL-3, and the cells were counted at the indicated times. The response of parental Ba/F3 cells in the presence or absence of mIL-3 is also shown. The results shown are the averages ± the standard deviations of triplicate cells. (C) Phosphorylation of STAT5A in parental Ba/F3 cells and Ba/F3 cells expressing subclone 5-1 with or without IL-3 stimulation. Stimulation with FCS had no effect on phosphorylation of STAT5A. STAT5A was immunoprecipitated by the anti-STAT5A antibody (R & D Systems) and blotted with the antiphosphotyrosine (P-Tyr) (Upstate Biotechnology Inc.) or with the anti-STAT5A antibody. (D) Long-term IL-3-independent proliferation of BaF/5A1*6 cells. BaF/5A1*6 cells were cultured in the absence of mIL-3 for the indicated periods and were counted. To exclude the possibility of autocrine growth, the growth curve of the parental Ba/F3 cells in the supernatant (BaF3 + 1*6 sup) of BaF/5A1*6 was also determined. The results shown are the averages ± the standard deviations of triplicate wells.
FIG. 2
A mutant STAT5B harboring mutations homologous to those in STAT5A1*6 induces IL-3-independent growth of Ba/F3 cells. Proliferation after transduction of the sequences for the wild-type STAT5A (5A WT) or STAT5B (5B WT) or mutated STAT5A (5A 1*6) STAT5B (5B 1*6) was examined with bulk populations of Ba/F3 cells after retrovirus-mediated transduction with these constructs. The infection efficiency in this experiment was 30 to 50%. Proliferation of parental Ba/F3 cells without IL-3 is also shown.
FIG. 3
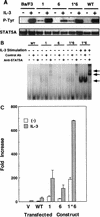
Characterization of the mutant STAT5As. (A) Tyrosine phosphorylation of STAT5A in Ba/F3 cells expressing the wild-type (WT) or the mutant STAT5As (1, 6, and 1*6) with (+) and without (−) IL-3 stimulation for 10 min. (B) DNA binding activities, without IL-3-stimulation, were examined by EMSA of the nuclear extracts derived from Ba/F3 cells expressing the wild-type STAT5A (WT) and the mutant STAT5As (1, 6, and 1*6). As a positive control, nuclear extracts were also prepared from Ba/F3 cells expressing the mutant STAT5A1*6 after stimulation with 10 ng of IL-3/ml for 10 min. The EMSA complexes were confirmed to contain STAT5A by supershift analysis with anti-STAT5A antibody. Arrows indicate the positions of the STAT5A bands as well as the supershifted complexes. Ab, antibody. (C) Transcriptional activation by the mutant STAT5As. Luciferase activities in the lysates of Ba/F3 cells transfected with the pMX vector (V), the sequence for the wild-type STAT5A (WT), and the sequence for the mutant STAT5As (1, 6, and 1*6) with or without IL-3 stimulation are shown. Transactivation observed in the control experiment (V) was derived from endogenous STAT5A and -5B. The results shown are the averages + the standard deviations of three independent experiments.
FIG. 4
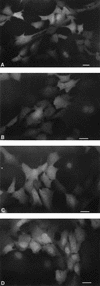
Nuclear localization of the mutant STAT5A. The fusion molecules STAT5A-EGFP (A), STAT5A1-EGFP (B), STAT5A6-EGFP (C), and STAT5A1*6-EGFP (D) were expressed in NIH 3T3 cells and observed under a fluorescence microscope. The photographs were taken with an automatic microscopic camera system (PM30; Olympus). Bars, 30 μm.
FIG. 5
Stability of the phosphorylated forms of the wild-type and the mutant STAT5As. Phosphorylation of the wild-type and the mutant STAT5As was examined in the transfectants (bulk population) expressing the wild-type STAT5A-Flag (wild type), STAT5A1-Flag (1), STAT5A6-Flag (6), and STAT5A1*6 (1*6). The cells were depleted of IL-3 for 12 h (−), stimulated with 10 ng of mIL-3/ml for 10 min, and cultured in the absence of mIL-3 for the indicated time periods, and lysates were prepared from the cells. The cell lysates were immunoprecipitated (IP) by the anti-Flag antibody and then blotted with 4G10 (α pY) or the anti-STAT5A antibody (α STAT5A). Arrows indicate positions of phosphorylated STAT5A-Flag.
FIG. 6
Cooperation of Raf and the STAT5 mutant in supporting IL-3-independent proliferation of Ba/F3 cells. (A) IL-3-independent proliferation of BR4 clones expressing the wild-type STAT5A (W1 and W2) and STAT5A1*6 (M1 through M6). BR4 clones which express both ΔRaf-ER and either the wild-type STAT5A or the mutant STAT5A1*6 were cultured in 96-well plates (103 cells/well) in RPMI 1640 medium supplemented with 0.5% BSA in the presence or absence of increasing concentrations of ICI for 3 days and subjected to an Alamar blue assay. Growth of parental BR4 cells (BR4) and the original Ba/F3 cells (P) are shown as controls. The results shown are the averages ± the standard deviations of triplicate wells (upper panel). O.D., optical density. To examine the expression level of STAT5A and ΔRaf-ER, total lysates (10 μg) from each clone were blotted with anti-STAT5A antibody and anti-ER antibody, respectively (lower panel). (B) Proliferation of BR4 cells expressing the wild-type (BR4/W) or the mutant STAT5A1*6 (BR4/M) in the absence of serum. Cells were cultured in 24-well plates (5 × 104/well) in RPMI 1640 medium containing 0.5% BSA, with (+) or without (−) 125 nM ICI, and were counted at the indicated times. The results shown are the averages ± the standard deviations of triplicate wells.
FIG. 7
Raf activation did not increase the DNA binding capability of either the wild-type or the mutant STAT5A. Nuclear extracts from the BR4 clones expressing the wild-type STAT5A (W1) and the mutant STAT5A (M6) and the parental BR4 cells were tested for DNA (PRE) binding by EMSA. The arrows indicate bands showing DNA binding and bands supershifted by the anti-STAT5 antibody. I, stimulated with ICI; 3, stimulated with IL-3. The M6 cells showed weak but significant DNA binding which was supershifted by the anti-STAT5A antibody but not by the control antibody (Ab).
Similar articles
- Constitutively active STAT5A and STAT5B in vitro and in vivo: mutation of STAT5 is not a frequent cause of leukemogenesis.
Yamada K, Ariyoshi K, Onishi M, Miyajima A, Hayakawa F, Towatari M, Saito H, Oka Y, Asano S, Nosaka T, Kitamura T. Yamada K, et al. Int J Hematol. 2000 Jan;71(1):46-54. Int J Hematol. 2000. PMID: 10729993 - Constitutive activation of STAT5 by a point mutation in the SH2 domain.
Ariyoshi K, Nosaka T, Yamada K, Onishi M, Oka Y, Miyajima A, Kitamura T. Ariyoshi K, et al. J Biol Chem. 2000 Aug 11;275(32):24407-13. doi: 10.1074/jbc.M909771199. J Biol Chem. 2000. PMID: 10823841 - Dominant negative mutants implicate STAT5 in myeloid cell proliferation and neutrophil differentiation.
Ilaria RL Jr, Hawley RG, Van Etten RA. Ilaria RL Jr, et al. Blood. 1999 Jun 15;93(12):4154-66. Blood. 1999. PMID: 10361113 - Stat5a and Stat5b: fraternal twins of signal transduction and transcriptional activation.
Grimley PM, Dong F, Rui H. Grimley PM, et al. Cytokine Growth Factor Rev. 1999 Jun;10(2):131-57. doi: 10.1016/s1359-6101(99)00011-8. Cytokine Growth Factor Rev. 1999. PMID: 10743504 Review. - The role of Stat5a and Stat5b in signaling by IL-2 family cytokines.
Lin JX, Leonard WJ. Lin JX, et al. Oncogene. 2000 May 15;19(21):2566-76. doi: 10.1038/sj.onc.1203523. Oncogene. 2000. PMID: 10851055 Review.
Cited by
- STAT5 is a key transcription factor for IL-3-mediated inhibition of RANKL-induced osteoclastogenesis.
Lee J, Seong S, Kim JH, Kim K, Kim I, Jeong BC, Nam KI, Kim KK, Hennighausen L, Kim N. Lee J, et al. Sci Rep. 2016 Aug 3;6:30977. doi: 10.1038/srep30977. Sci Rep. 2016. PMID: 27485735 Free PMC article. - Src family kinases interfere with dimerization of STAT5A through a phosphotyrosine-SH2 domain interaction.
Fahrenkamp D, de Leur HS, Küster A, Chatain N, Müller-Newen G. Fahrenkamp D, et al. Cell Commun Signal. 2015 Feb 15;13:10. doi: 10.1186/s12964-014-0081-7. Cell Commun Signal. 2015. PMID: 25885255 Free PMC article. - Strategies to enhance CAR-T persistence.
Liu Y, An L, Huang R, Xiong J, Yang H, Wang X, Zhang X. Liu Y, et al. Biomark Res. 2022 Nov 23;10(1):86. doi: 10.1186/s40364-022-00434-9. Biomark Res. 2022. PMID: 36419115 Free PMC article. Review. - Productive parvovirus B19 infection of primary human erythroid progenitor cells at hypoxia is regulated by STAT5A and MEK signaling but not HIFα.
Chen AY, Kleiboeker S, Qiu J. Chen AY, et al. PLoS Pathog. 2011 Jun;7(6):e1002088. doi: 10.1371/journal.ppat.1002088. Epub 2011 Jun 16. PLoS Pathog. 2011. PMID: 21698228 Free PMC article. - Prostaglandin E2-EP2/EP4 signaling induces immunosuppression in human cancer by impairing bioenergetics and ribosome biogenesis in immune cells.
Punyawatthananukool S, Matsuura R, Wongchang T, Katsurada N, Tsuruyama T, Tajima M, Enomoto Y, Kitamura T, Kawashima M, Toi M, Yamanoi K, Hamanishi J, Hisamori S, Obama K, Charoensawan V, Thumkeo D, Narumiya S. Punyawatthananukool S, et al. Nat Commun. 2024 Nov 1;15(1):9464. doi: 10.1038/s41467-024-53706-3. Nat Commun. 2024. PMID: 39487111 Free PMC article.
References
- Ahmed S A, Gogal R, Jr, Walsh J E. A new rapid and simple non-radioactive assay to monitor and determine the proliferation of lymphocytes: an alternative to [3H]thymidine incorporation assay. J Immunol Methods. 1994;170:211–224. - PubMed
- Azam M, Lee C, Strehlow I, Schindler C. Functionally distinct isoforms of STAT5 are generated by protein processing. Immunity. 1997;6:691–701. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous