Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3 - PubMed (original) (raw)
Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3
H Niwa et al. Genes Dev. 1998.
Abstract
The propagation of embryonic stem (ES) cells in an undifferentiated pluripotent state is dependent on leukemia inhibitory factor (LIF) or related cytokines. These factors act through receptor complexes containing the signal transducer gp130. The downstream mechanisms that lead to ES cell self-renewal have not been delineated, however. In this study, chimeric receptors were introduced into ES cells. Biochemical and functional studies of transfected cells demonstrated a requirement for engagement and activation of the latent trancription factor STAT3. Detailed mutational analyses unexpectedly revealed that the four STAT3 docking sites in gp130 are not functionally equivalent. The role of STAT3 was then investigated using the dominant interfering mutant, STAT3F. ES cells that expressed this molecule constitutively could not be isolated. An episomal supertransfection strategy was therefore used to enable the consequences of STAT3F expression to be examined. In addition, an inducible STAT3F transgene was generated. In both cases, expression of STAT3F in ES cells growing in the presence of LIF specifically abrogated self-renewal and promoted differentiation. These complementary approaches establish that STAT3 plays a central role in the maintenance of the pluripotential stem cell phenotype. This contrasts with the involvement of STAT3 in the induction of differentiation in somatic cell types. Cell type-specific interpretation of STAT3 activation thus appears to be pivotal to the diverse developmental effects of the LIF family of cytokines. Identification of STAT3 as a key transcriptional determinant of ES cell self-renewal represents a first step in the molecular characterization of pluripotency.
Figures
Figure 1
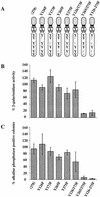
ES cell self-renewal and induction of STAT DNA-binding activity mediated by G-CSF-R wild-type, truncated, and chimeric cytokine receptors. (A) Efficiency of clonal stem cell renewal in response to G-CSF measured by formation of alkaline phosphatase-positive colonies. (Light gray bars) −G-CSF; (dark gray bars) +G-CSF. Data are mean ±
s.e.m.
of triplicate determinations on single representative clones normalized to response to IL-6/sIL-6R. (B) Induction of STAT DNA binding by IL-6/sIL-6R and G-CSF determined by electophoretic mobility-shift assay. Cells were untreated or stimulated for 30 min with IL-6/sIL-6R or G-CSF (30 ng/ml). Nuclear extracts were prepared and assayed for SIE binding. Note the absence of detectable STAT1/STAT3 heterodimer complex on stimulation of full-length G-CSF-R.
Figure 1
ES cell self-renewal and induction of STAT DNA-binding activity mediated by G-CSF-R wild-type, truncated, and chimeric cytokine receptors. (A) Efficiency of clonal stem cell renewal in response to G-CSF measured by formation of alkaline phosphatase-positive colonies. (Light gray bars) −G-CSF; (dark gray bars) +G-CSF. Data are mean ±
s.e.m.
of triplicate determinations on single representative clones normalized to response to IL-6/sIL-6R. (B) Induction of STAT DNA binding by IL-6/sIL-6R and G-CSF determined by electophoretic mobility-shift assay. Cells were untreated or stimulated for 30 min with IL-6/sIL-6R or G-CSF (30 ng/ml). Nuclear extracts were prepared and assayed for SIE binding. Note the absence of detectable STAT1/STAT3 heterodimer complex on stimulation of full-length G-CSF-R.
Figure 2
Effect of mutating STAT3 interaction sites in gp130 on ES cell self-renewal and induction of STAT3 DNA-binding activity. (A) Schematic of the various chimeric receptors indicating the tyrosine–phenylalanine substitutions introduced into the wild-type (278) gp130 cytoplasmic domain. Numbering commences with the first residue of the 278-amino-acid intracellular domain of mouse gp130. The phenylalanine (F) for tyrosine (Y) substitutions in the four STAT3 docking sites are indicated. The additional three tyrosines do not interact with STAT3 (Stahl et al. 1995). (B) Stem cell renewal mediated by chimeric receptors in response to G-CSF measured by β-galactosidase expression from the Oct-4 locus. Data are mean ±
s.e.m.
for duplicate determinations on three independent clones normalized relative to response to IL-6/sIL-6R. (C) Efficiency of clonal stem cell renewal mediated by chimeric receptors in response to G-CSF measured by formation of alkaline phosphatase positive colonies. Data are mean ±
s.e.m.
for duplicate assays on three independent clones normalized relative to response to IL-6/sIL-6R. (D) Electrophoretic mobility-shift assay of induced STAT3 DNA binding. Transfected clones were left untreated (lane 1) or stimulated for 30 min with IL-6/sIL-6R (lane 2) or with G-CSF at 30 ng/ml (lane 3) or 3 ng/ml (lane 4). Nuclear extracts were assayed for SIE binding. (E) Immunoblot of STAT3 and ERK phosphorylation induced by G-CSF stimulation of chimeric receptors. Transfected clones were left untreated (lane 1) or were stimulated for 20 min with IL-6/sIL-6R (lane 2) or with G-CSF (lane 3). Immunoblots of cell lysates were probed sequentially with antibodies specific for the active phosphorylated forms of ERK and STAT3.
Figure 2
Effect of mutating STAT3 interaction sites in gp130 on ES cell self-renewal and induction of STAT3 DNA-binding activity. (A) Schematic of the various chimeric receptors indicating the tyrosine–phenylalanine substitutions introduced into the wild-type (278) gp130 cytoplasmic domain. Numbering commences with the first residue of the 278-amino-acid intracellular domain of mouse gp130. The phenylalanine (F) for tyrosine (Y) substitutions in the four STAT3 docking sites are indicated. The additional three tyrosines do not interact with STAT3 (Stahl et al. 1995). (B) Stem cell renewal mediated by chimeric receptors in response to G-CSF measured by β-galactosidase expression from the Oct-4 locus. Data are mean ±
s.e.m.
for duplicate determinations on three independent clones normalized relative to response to IL-6/sIL-6R. (C) Efficiency of clonal stem cell renewal mediated by chimeric receptors in response to G-CSF measured by formation of alkaline phosphatase positive colonies. Data are mean ±
s.e.m.
for duplicate assays on three independent clones normalized relative to response to IL-6/sIL-6R. (D) Electrophoretic mobility-shift assay of induced STAT3 DNA binding. Transfected clones were left untreated (lane 1) or stimulated for 30 min with IL-6/sIL-6R (lane 2) or with G-CSF at 30 ng/ml (lane 3) or 3 ng/ml (lane 4). Nuclear extracts were assayed for SIE binding. (E) Immunoblot of STAT3 and ERK phosphorylation induced by G-CSF stimulation of chimeric receptors. Transfected clones were left untreated (lane 1) or were stimulated for 20 min with IL-6/sIL-6R (lane 2) or with G-CSF (lane 3). Immunoblots of cell lysates were probed sequentially with antibodies specific for the active phosphorylated forms of ERK and STAT3.
Figure 3
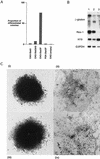
Induction of differentiation by expression of STAT3F in MG1.19 ES cells. (A) Proportion of differentiated colonies in LIF-supplemented medium resulting from supertransfection of STAT3, antisense STAT3, and STAT3F expression vectors. Colonies were fixed and stained with Leishman’s reagent after 8 days of selection, and the numbers of stem cell colonies and differentiated colonies were scored. (B) Marker gene expression in STAT3F supertransfectants. Expression of marker genes in pools of MG1.19 cells supertransfected with STAT3 (lane 1), STAT3 antisense (lane 2), and STAT3F (lane 3) expression vectors. Total RNA was prepared after 8 days of selection in LIF-supplemented medium, and 5-μg aliquots were analyzed by filter hybridization with β-globin, Rex-1, H19, and G3PDH probes. The β-globin probe detects all transgene mRNA species generated from pHPCAG, including an alternatively spliced product from the antisense contruct. (C) Photomicrographs of representative colonies 8 days after supertransfection with (i) STAT3, (ii) STAT3F, and (iii) empty expression vectors and selection in the presence of LIF, or (iv) induction of differentiation by culture in the absence of LIF for 8 days.
Figure 4
Cosupertransfection of STAT3F with wild-type STAT expression vectors. Proportions of undifferentiated stem cell colonies generated after cosupertransfection of MG1.19 ES cells with 10 μg of pBPCAGGS–STAT3F plus 10 μg of pHPCAG vector containing stuffer (control), STAT3, STAT1, or STAT4 inserts. After 8 days of selection with 80 μg/ml of hygromycin B plus 20 μg/ml of blasticidin S, colonies were fixed and stained with Leishman’s reagent.
Figure 5
Generation of an inducible STAT3F transgene integration by supertargeting. (A) Schematic of supertargeting strategy for introduction of STAT3F into a tetracycline-regulatable expression site. ZHTc6 ES cells contain a tetracycline-regulated transgene comprising the hCMV*-1 promoter (Gossen and Bujard 1992), β-globin second intron, Oct-4 open reading frame (Okazawa et al. 1991), and IRESβgeopA selection marker (Mountford et al. 1994). Homologous recombination can be used to replace the Oct-4 sequence (supertargeting). Use of a truncated selection marker in the targeting vector facilitates the isolation of homologous recombinants. ZHTc6 cells were electroporated with the STAT3F–SuperKO vector and selected in G418 in the presence of tetracycline. G418-resistant clones were duplicated and screened for sensitivity against gancyclovir to enrich further for homologous recombinants. The option of excising the _loxP_-flanked MC1tk cassette by transient expression of Cre recombinase was not pursued. (B) Diagnosis of the supertargeting event in Gs ES cells. Gancyclovir-sensitive (Gs; lanes 1–4) and -resistant (Gr; lane 5) clones were analyzed by Southern hybridization. A 3.2-kb _Sac_I fragment was detected with a probe from the 5′ end of lacZ in the Gs samples, indicative of the correct replacement of the Oct-4 cDNA sequence with STAT3F sequence. The Gr clone retained the 4.8-kb fragment diagnostic for the original Oct-4 transgene integration in ZHTc6 cells.
Figure 5
Generation of an inducible STAT3F transgene integration by supertargeting. (A) Schematic of supertargeting strategy for introduction of STAT3F into a tetracycline-regulatable expression site. ZHTc6 ES cells contain a tetracycline-regulated transgene comprising the hCMV*-1 promoter (Gossen and Bujard 1992), β-globin second intron, Oct-4 open reading frame (Okazawa et al. 1991), and IRESβgeopA selection marker (Mountford et al. 1994). Homologous recombination can be used to replace the Oct-4 sequence (supertargeting). Use of a truncated selection marker in the targeting vector facilitates the isolation of homologous recombinants. ZHTc6 cells were electroporated with the STAT3F–SuperKO vector and selected in G418 in the presence of tetracycline. G418-resistant clones were duplicated and screened for sensitivity against gancyclovir to enrich further for homologous recombinants. The option of excising the _loxP_-flanked MC1tk cassette by transient expression of Cre recombinase was not pursued. (B) Diagnosis of the supertargeting event in Gs ES cells. Gancyclovir-sensitive (Gs; lanes 1–4) and -resistant (Gr; lane 5) clones were analyzed by Southern hybridization. A 3.2-kb _Sac_I fragment was detected with a probe from the 5′ end of lacZ in the Gs samples, indicative of the correct replacement of the Oct-4 cDNA sequence with STAT3F sequence. The Gr clone retained the 4.8-kb fragment diagnostic for the original Oct-4 transgene integration in ZHTc6 cells.
Figure 6
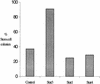
Induced expression of STAT3F causes ES cell differentiation and inhibits STAT3 activation. (A) Differentiation of Gs ES cells induced by withdrawal of tetracycline. Gs ES cells grown up in the presence of tetracycline were plated at clonal density (500 cells/60-mm dish) in LIF-supplemented medium in the presence or absence of tetracycline (1 μg/ml). After 6 days, colonies were fixed and stained with Leishman’s reagent. The histogram records the proportions of differentiated colonies for three independent clones, Gs1 (solid bars), Gs2 (hatched bars), and Gs3 (shaded bars). (B) Dose response curve of Gs2 cell differentiation. Gs2 ES cells were cultured as above in the presence of the indicated concentrations of tetracycline, then fixed, stained, and scored. (C) Photomicrographs of uninduced and induced Gs2 ES cells. Representative colonies of Gs2 cells cultured for 6 days in LIF-supplemented medium in the presence (+Tc) or absence (−Tc) of tetracycline (1 μg/ml) and then fixed and stained with Leishman’s reagent. (D) Mobility retardation assay of STAT3 DNA-binding activity in noninduced and induced Gs2 cells. Gs2 ES cells were cultured for 72 hr in the presence or absence of tetracycline. IL-6/sIL-6R was withdrawn for the final 24 hr, then restored for the indicated times. Nuclear extracts were prepared and assayed as described for SIE DNA-binding activity. (E) Quantitation of STAT3 SIE binding by PhosphorImager. (Shaded bars) +Tc; (open bars) −Tc.
Figure 6
Induced expression of STAT3F causes ES cell differentiation and inhibits STAT3 activation. (A) Differentiation of Gs ES cells induced by withdrawal of tetracycline. Gs ES cells grown up in the presence of tetracycline were plated at clonal density (500 cells/60-mm dish) in LIF-supplemented medium in the presence or absence of tetracycline (1 μg/ml). After 6 days, colonies were fixed and stained with Leishman’s reagent. The histogram records the proportions of differentiated colonies for three independent clones, Gs1 (solid bars), Gs2 (hatched bars), and Gs3 (shaded bars). (B) Dose response curve of Gs2 cell differentiation. Gs2 ES cells were cultured as above in the presence of the indicated concentrations of tetracycline, then fixed, stained, and scored. (C) Photomicrographs of uninduced and induced Gs2 ES cells. Representative colonies of Gs2 cells cultured for 6 days in LIF-supplemented medium in the presence (+Tc) or absence (−Tc) of tetracycline (1 μg/ml) and then fixed and stained with Leishman’s reagent. (D) Mobility retardation assay of STAT3 DNA-binding activity in noninduced and induced Gs2 cells. Gs2 ES cells were cultured for 72 hr in the presence or absence of tetracycline. IL-6/sIL-6R was withdrawn for the final 24 hr, then restored for the indicated times. Nuclear extracts were prepared and assayed as described for SIE DNA-binding activity. (E) Quantitation of STAT3 SIE binding by PhosphorImager. (Shaded bars) +Tc; (open bars) −Tc.
Figure 6
Induced expression of STAT3F causes ES cell differentiation and inhibits STAT3 activation. (A) Differentiation of Gs ES cells induced by withdrawal of tetracycline. Gs ES cells grown up in the presence of tetracycline were plated at clonal density (500 cells/60-mm dish) in LIF-supplemented medium in the presence or absence of tetracycline (1 μg/ml). After 6 days, colonies were fixed and stained with Leishman’s reagent. The histogram records the proportions of differentiated colonies for three independent clones, Gs1 (solid bars), Gs2 (hatched bars), and Gs3 (shaded bars). (B) Dose response curve of Gs2 cell differentiation. Gs2 ES cells were cultured as above in the presence of the indicated concentrations of tetracycline, then fixed, stained, and scored. (C) Photomicrographs of uninduced and induced Gs2 ES cells. Representative colonies of Gs2 cells cultured for 6 days in LIF-supplemented medium in the presence (+Tc) or absence (−Tc) of tetracycline (1 μg/ml) and then fixed and stained with Leishman’s reagent. (D) Mobility retardation assay of STAT3 DNA-binding activity in noninduced and induced Gs2 cells. Gs2 ES cells were cultured for 72 hr in the presence or absence of tetracycline. IL-6/sIL-6R was withdrawn for the final 24 hr, then restored for the indicated times. Nuclear extracts were prepared and assayed as described for SIE DNA-binding activity. (E) Quantitation of STAT3 SIE binding by PhosphorImager. (Shaded bars) +Tc; (open bars) −Tc.
Similar articles
- STAT3 is dispensable for maintenance of self-renewal in nonhuman primate embryonic stem cells.
Sumi T, Fujimoto Y, Nakatsuji N, Suemori H. Sumi T, et al. Stem Cells. 2004;22(5):861-72. doi: 10.1634/stemcells.22-5-861. Stem Cells. 2004. PMID: 15342949 - Essential role of STAT3 for embryonic stem cell pluripotency.
Raz R, Lee CK, Cannizzaro LA, d'Eustachio P, Levy DE. Raz R, et al. Proc Natl Acad Sci U S A. 1999 Mar 16;96(6):2846-51. doi: 10.1073/pnas.96.6.2846. Proc Natl Acad Sci U S A. 1999. PMID: 10077599 Free PMC article. - Paracrine induction of stem cell renewal by LIF-deficient cells: a new ES cell regulatory pathway.
Dani C, Chambers I, Johnstone S, Robertson M, Ebrahimi B, Saito M, Taga T, Li M, Burdon T, Nichols J, Smith A. Dani C, et al. Dev Biol. 1998 Nov 1;203(1):149-62. doi: 10.1006/dbio.1998.9026. Dev Biol. 1998. PMID: 9806780 - Animal embryonic stem (ES) cells: self-renewal, pluripotency, transgenesis and nuclear transfer.
Saito S, Liu B, Yokoyama K. Saito S, et al. Hum Cell. 2004 Sep;17(3):107-15. doi: 10.1111/j.1749-0774.2004.tb00026.x. Hum Cell. 2004. PMID: 15859155 Review. - Cytokine signalling in embryonic stem cells.
Kristensen DM, Kalisz M, Nielsen JH. Kristensen DM, et al. APMIS. 2005 Nov-Dec;113(11-12):756-72. doi: 10.1111/j.1600-0463.2005.apm_391.x. APMIS. 2005. PMID: 16480448 Review.
Cited by
- Wnt pathway regulation of embryonic stem cell self-renewal.
Merrill BJ. Merrill BJ. Cold Spring Harb Perspect Biol. 2012 Sep 1;4(9):a007971. doi: 10.1101/cshperspect.a007971. Cold Spring Harb Perspect Biol. 2012. PMID: 22952393 Free PMC article. Review. - METTLing in Stem Cell and Cancer Biology.
Tooley JG, Catlin JP, Tooley CES. Tooley JG, et al. Stem Cell Rev Rep. 2023 Jan;19(1):76-91. doi: 10.1007/s12015-022-10444-7. Epub 2022 Sep 12. Stem Cell Rev Rep. 2023. PMID: 36094754 Free PMC article. Review. - Small molecules, big roles -- the chemical manipulation of stem cell fate and somatic cell reprogramming.
Zhang Y, Li W, Laurent T, Ding S. Zhang Y, et al. J Cell Sci. 2012 Dec 1;125(Pt 23):5609-20. doi: 10.1242/jcs.096032. J Cell Sci. 2012. PMID: 23420199 Free PMC article. Review. - Changes in the phosphorylation of nucleotide metabolism‑associated proteins by leukemia inhibitory factor in mouse embryonic stem cells.
Song HR, Kim HK, Kim SG, Lim HJ, Kim HY, Han MK. Song HR, et al. Mol Med Rep. 2021 Jun;23(6):431. doi: 10.3892/mmr.2021.12070. Epub 2021 Apr 13. Mol Med Rep. 2021. PMID: 33846773 Free PMC article. - A novel role for P2X7 receptor signalling in the survival of mouse embryonic stem cells.
Thompson BA, Storm MP, Hewinson J, Hogg S, Welham MJ, MacKenzie AB. Thompson BA, et al. Cell Signal. 2012 Mar;24(3):770-8. doi: 10.1016/j.cellsig.2011.11.012. Epub 2011 Nov 18. Cell Signal. 2012. PMID: 22120528 Free PMC article.
References
- Argetsinger LS, Hsu GW, Myers MG, Billestrup N, White MF, Carter-Su C. Growth hormone, interferon-γ, and leukemia inhibitory factor promoted tyrosyl phosphorylation of insulin receptor substrate-1. J Biol Chem. 1995;270:14685–14692. - PubMed
- Baumann H, Wong GG. Hepatocyte-stimulating factor-lll shares structural and functional identity with leukemia-inhibitory factor. J Immunol. 1989;143:1163–1167. - PubMed
- Baumann H, Gearing D, Ziegler S. Signaling by the cytoplasmic domain of hematopoietin receptors involves two distinguishable mechanisms in hepatic cells. J Biol Chem. 1994a;269:16297–16304. - PubMed
- Baumann H, Symes AJ, Comeau MR, Morella KK, Wang Y, Friend D, Ziegler SF, Fink JS, Gearing DP. Multiple regions within the cytoplasmic domains of the leukemia inhibitory factor receptor and gp130 cooperate in signal transduction in hepatic and neuronal cells. Mol Cell Biol. 1994b;14:138–146. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous