The CC chemokine 6Ckine binds the CXC chemokine receptor CXCR3 - PubMed (original) (raw)
The CC chemokine 6Ckine binds the CXC chemokine receptor CXCR3
H Soto et al. Proc Natl Acad Sci U S A. 1998.
Abstract
We cloned the mouse homologue of the chemokine receptor CXCR3, which is located in mouse chromosome X. We screened a large panel of chemokines for their ability to induce a calcium flux in mouse CXCR3-transfected cells and identified a new ligand for this receptor, the recently reported CC chemokine 6Ckine. This represents an example of a CC chemokine, which binds to a CXC chemokine receptor. Like other ligands of this receptor, 6Ckine has angiostatic properties. 6Ckine is known to chemoattract T cells. In line with this, CXCR3 is expressed preferentially in Th1 cells and in lymphoid organs of the IL-10(-/-) mouse that develops chronic colitis. Its ability to attract T cells as well as its angiostatic properties suggest that 6Ckine may be an effective anti-tumor agent.
Figures
Figure 1
Aligment of the mCXCR3 predicted amino acid sequence with hCXCR3. The arrowhead indicates potential N-linked glycosilation sites and the horizontal lines the putative TMD (TMD1–TMD7). The alignment was generated by using
clustalw
(17). The sequence data are available from European Molecular Biology Laboratory (EMBL)/GenBank under accession no. AF045146.
Figure 2
Distribution of mRNA of mCXCR3. (A) Multiple tissue Northern blot was hybridized with a full-length mCXCR3 cDNA probe, and two bands of 1.7 and 3.6 kb were detected in lung, spleen, and heart. (B) Southern blot analysis of different cDNA libraries: Th1 1 wk, naive cells from spleen were polarized for 7 days with IFNγ and anti-IL-4; Th2 1 wk, naive cells were polarized for 7 days with IL-4 and anti-IFNγ. Th1 3 wk: naive cells from Rag1−/− × DO-11.10-transgenic mice were stimulated with ovalbumin-(323–339) peptide in the presence of irradiated splenic APC and IL-12 and anti-IL-4 for 3 wk. Th2 3 wk: naive cells from transgenic mice (similar to Th1 3 wk) were stimulated with anti-IL-12 plus IL-4 for 3 wk; IL-10−/− mice Peyer’s patches; Peyer’s patches; IL-10−/− mice mesenteric lymph nodes; mouse mesenteric lymph nodes; B cells derived from spleen; and mouse endothelial cells. (C) CD4+NK1.1+-activated T cells; αβTCR+CD4−CD8− resting T cells; αβTCR+CD4−CD8− -activated T cells; pro-T-resting cells; and pro-T-activated cells.
Figure 3
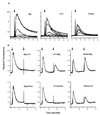
Calcium flux and desensitization analysis. (A) Calcium mobilization and dose response of Mig, IP-10, and 6Ckine with mouse CXCR3 stable-transfected HEK293 cells. Arrow indicates the time point of addition of the indicated ligands. Each chemokine was loaded from 1 nmol to 1 μmol in concentration. (B) Cross desensitization among Mig, IP-10, and 6Ckine. Arrows indicate the first and the second addition of the indicated chemokines. The chemokines were loaded at 1 μmol in concentration to induce a maximum calcium mobilization response.
Figure 4
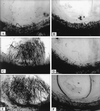
Angiostatic activity of 6Ckine. Angiogenic response in the rat corneal micropocket assay to bFGF (C) (5 nM) and VEGF (E) (5 nM) were inhibited by 6Ckine (10 nM) (D and F, respectively). Control (A) was DMEM with 0.1% BSA alone or 6Ckine alone (B).
Figure 5
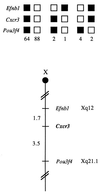
Cxcr3 maps to the central region of the mouse chromosome X. Cxcr3 was placed on the mouse chromosome X by interspecific backcross analysis. The segregation patterns of Cxcr3 and flanking genes in 161 backcross animals that were typed (see text). Each column represents the chromosome identified in the backcross progeny that was inherited from the (C57BL/6J × M. spretus)F1 parent. The shaded boxes represent the presence of a C57BL/6J allele, and white boxes represent the presence of a M. spretus allele. The number of offspring inheriting each type of chromosome is listed at the bottom of each column. A partial X chromosome-linkage map showing the location of Cxcr3 in relation to linked genes is shown at the bottom of the figure. Recombination distances between loci in centimorgans are shown to the left of the chromosome and the positions of the loci in human chromosomes, where known, are show to the right. References for the human map positions of loci cited in this study can be obtained from Genome Database (GDB), a computerized database of human linkage information maintained by the William H. Welcher Medical Library of The Johns Hopkins University (Baltimore, MD).
Similar articles
- Cutting edge: species specificity of the CC chemokine 6Ckine signaling through the CXC chemokine receptor CXCR3: human 6Ckine is not a ligand for the human or mouse CXCR3 receptors.
Jenh CH, Cox MA, Kaminski H, Zhang M, Byrnes H, Fine J, Lundell D, Chou CC, Narula SK, Zavodny PJ. Jenh CH, et al. J Immunol. 1999 Apr 1;162(7):3765-9. J Immunol. 1999. PMID: 10201891 - Cutting edge: activity of human adult microglia in response to CC chemokine ligand 21.
Dijkstra IM, Hulshof S, van der Valk P, Boddeke HW, Biber K. Dijkstra IM, et al. J Immunol. 2004 Mar 1;172(5):2744-7. doi: 10.4049/jimmunol.172.5.2744. J Immunol. 2004. PMID: 14978072 - Antitumor effects of the mouse chemokine 6Ckine/SLC through angiostatic and immunological mechanisms.
Vicari AP, Ait-Yahia S, Chemin K, Mueller A, Zlotnik A, Caux C. Vicari AP, et al. J Immunol. 2000 Aug 15;165(4):1992-2000. doi: 10.4049/jimmunol.165.4.1992. J Immunol. 2000. PMID: 10925282 - Novel lymphocyte-specific CC chemokines and their receptors.
Yoshie O, Imai T, Nomiyama H. Yoshie O, et al. J Leukoc Biol. 1997 Nov;62(5):634-44. doi: 10.1002/jlb.62.5.634. J Leukoc Biol. 1997. PMID: 9365118 Review. - Chemokine receptor CXCR3: an unexpected enigma.
Liu L, Callahan MK, Huang D, Ransohoff RM. Liu L, et al. Curr Top Dev Biol. 2005;68:149-81. doi: 10.1016/S0070-2153(05)68006-4. Curr Top Dev Biol. 2005. PMID: 16124999 Review.
Cited by
- Systematic classification of vertebrate chemokines based on conserved synteny and evolutionary history.
Nomiyama H, Osada N, Yoshie O. Nomiyama H, et al. Genes Cells. 2013 Jan;18(1):1-16. doi: 10.1111/gtc.12013. Epub 2012 Nov 12. Genes Cells. 2013. PMID: 23145839 Free PMC article. Review. - Expression of chemokine receptors in vernal keratoconjunctivitis.
Abu El-Asrar AM, Struyf S, Al-Mosallam AA, Missotten L, Van Damme J, Geboes K. Abu El-Asrar AM, et al. Br J Ophthalmol. 2001 Nov;85(11):1357-61. doi: 10.1136/bjo.85.11.1357. Br J Ophthalmol. 2001. PMID: 11673306 Free PMC article. - Alternative medicines as emerging therapies for inflammatory bowel diseases.
Singh UP, Singh NP, Busbee B, Guan H, Singh B, Price RL, Taub DD, Mishra MK, Nagarkatti M, Nagarkatti PS. Singh UP, et al. Int Rev Immunol. 2012 Feb;31(1):66-84. doi: 10.3109/08830185.2011.642909. Int Rev Immunol. 2012. PMID: 22251008 Free PMC article. Review. - The CC chemokine thymus-derived chemotactic agent 4 (TCA-4, secondary lymphoid tissue chemokine, 6Ckine, exodus-2) triggers lymphocyte function-associated antigen 1-mediated arrest of rolling T lymphocytes in peripheral lymph node high endothelial venules.
Stein JV, Rot A, Luo Y, Narasimhaswamy M, Nakano H, Gunn MD, Matsuzawa A, Quackenbush EJ, Dorf ME, von Andrian UH. Stein JV, et al. J Exp Med. 2000 Jan 3;191(1):61-76. doi: 10.1084/jem.191.1.61. J Exp Med. 2000. PMID: 10620605 Free PMC article. - Cell cycle-dependent expression of CXC chemokine receptor 3 by endothelial cells mediates angiostatic activity.
Romagnani P, Annunziato F, Lasagni L, Lazzeri E, Beltrame C, Francalanci M, Uguccioni M, Galli G, Cosmi L, Maurenzig L, Baggiolini M, Maggi E, Romagnani S, Serio M. Romagnani P, et al. J Clin Invest. 2001 Jan;107(1):53-63. doi: 10.1172/JCI9775. J Clin Invest. 2001. PMID: 11134180 Free PMC article.
References
- Rollins B J. Blood. 1997;90:909–928. - PubMed
- Baggiolini M, Dewald B, Moser B. Annu Rev Immunol. 1997;15:675–705. - PubMed
- Kelner G, Kennedy J, Bacon K, Kleyensteuber S, Largaespada D, Jenkins N, Copeland N, Bazan J F, Moore K, Schall T J, et al. Science. 1994;266:1395–1399. - PubMed
- Bazan J F, Bacon K B, Hardiman G, Wang W, Soo K, Rossi D, Greavbes D R, Zlotnik A, Schall T J. Nature (London) 1997;385:640–664. - PubMed
- Hedrick J A, Zlotnik A. J Immunol. 1997;159:1589–1593. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases