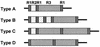Variants of the 3' region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases - PubMed (original) (raw)
Variants of the 3' region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases
Y Yamaoka et al. J Clin Microbiol. 1998 Aug.
Abstract
The CagA protein of Helicobacter pylori is an immunogenic antigen of variable size and unknown function that has been associated with increased virulence as well as two mutually exclusive diseases, duodenal ulcer and gastric carcinoma. The 3' region of the cagA gene contains repeated sequences. To determine whether there are structural changes in the 3' region of cagA that predict outcome of H. pylori infection, we examined 155 cagA gene-positive H. pylori isolates from Japanese patients including 50 patients with simple gastritis, 40 with gastric ulcer, 35 with duodenal ulcer, and 30 with gastric cancer. The 3' region of the cagA gene was amplified by PCR followed by sequencing. CagA proteins were detected by immunoblotting using a polyclonal antibody against recombinant CagA. One hundred forty-five strains yielded PCR products of 642 to 651 bp; 10 strains had products of 756 to 813 bp. The sequence of the 3' region of the cagA gene in Japan differs markedly from the primary sequence of cagA genes from Western isolates. Sequence analysis of the PCR products showed four types of primary gene structure (designated types A, B, C, and D) depending on the type and number of repeats. Six of the seven type C strains were found in patients with gastric cancer (P < 0.01 in comparison to noncancer patients). Comparison of type A and type C strains from patients with gastric cancer showed that type C was associated with higher levels of CagA antibody and more severe degrees of atrophy. Differences in cagA genotype may be useful for molecular epidemiology and may provide a marker for differences in virulence among cagA-positive H. pylori strains.
Figures
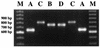
FIG. 1
Analysis of the 3′ region of the cagA gene by PCR. PCR products from a representative group of strains are shown. The sizes of the DNA fragments were confirmed after sequencing of the PCR products. Fragments A had sizes ranging from 642 to 651 bp. Sequencing showed that the primary structure of the cagA gene of two strains with 756-bp PCR fragments was different, and these strains were designated B and D. Fragments C were 810-bp products. Lanes M, molecular size markers.
FIG. 2
Primary-structure variants of the 3′ region of the cagA gene in a Japanese population. The fragments are not represented on a proportional scale.
FIG. 3
Sequence analyses of type A and type C cagA genes. (A) Alignment of the deduced amino acid sequences of type C strains, in the region corresponding to amino acid residues 892 to 969 of the H. pylori ATCC 53726 CagA product (GenBank accession no. L11714). Strain JK14 was from a patient with gastric ulcer, and the remaining six strains were from patients with gastric cancer. (B) Comparison of the deduced amino acid sequences of the R1, R2, and R3 regions of type A and type C strains with the same region of the gene product of a reference strain. The reference strains ATCC 43526, G39, ATCC 53726, and CCUG17874 (GenBank accession no. AB003397, X70038, L11714, and X70039, respectively) were used for sequence comparison. Consensus sequences among the cagA genes from H. pylori isolates from patients with chronic gastritis (CG), gastric ulcer (GU), duodenal ulcer (DU), and gastric cancer (GC) were separately determined among type A strains.
FIG. 4
Western blot analysis of CagA proteins. A1 through A4, B1 and B2, C1 through C3, and D represent examples of strains with 3′ region structures that characterize cagA genes of types A to D. The molecular weights of proteins were determined in comparison to molecular weight standards run on the same gel (Kaleidoscope prestained standards) and by comparison with the size of CagA from the reference strain CCUG17874 (128K) (lane CC). The molecular weights of the CagA proteins are indicated at the bottom.
Comment in
- Clarifications regarding the 3' repeat region of the cagA gene in Helicobacter pylori and clinical outcome.
Yamaoka Y, Graham DY. Yamaoka Y, et al. J Clin Microbiol. 2001 Jun;39(6):2369-70. doi: 10.1128/JCM.39.6.2369-2370.2001. J Clin Microbiol. 2001. PMID: 11414245 Free PMC article. No abstract available.
Similar articles
- A new subtype of 3' region of cagA gene in Helicobacter pylori strains isolated from Zhejiang Province in China.
Tao R, Fang PC, Liu HY, Jiang YS, Chen J. Tao R, et al. World J Gastroenterol. 2004 Nov 15;10(22):3284-8. doi: 10.3748/wjg.v10.i22.3284. World J Gastroenterol. 2004. PMID: 15484301 Free PMC article. - Relationship between the diversity of the cagA gene of Helicobacter pylori and gastric cancer in Okinawa, Japan.
Satomi S, Yamakawa A, Matsunaga S, Masaki R, Inagaki T, Okuda T, Suto H, Ito Y, Yamazaki Y, Kuriyama M, Keida Y, Kutsumi H, Azuma T. Satomi S, et al. J Gastroenterol. 2006 Jul;41(7):668-73. doi: 10.1007/s00535-006-1838-6. J Gastroenterol. 2006. PMID: 16933004 - H. pylori and cagA: relationships with gastric cancer, duodenal ulcer, and reflux esophagitis and its complications.
Graham DY, Yamaoka Y. Graham DY, et al. Helicobacter. 1998 Sep;3(3):145-51. doi: 10.1046/j.1523-5378.1998.08031.x. Helicobacter. 1998. PMID: 9731983 Review. - Helicobacter pylori typing as a tool for tracking human migration.
Yamaoka Y. Yamaoka Y. Clin Microbiol Infect. 2009 Sep;15(9):829-34. doi: 10.1111/j.1469-0691.2009.02967.x. Clin Microbiol Infect. 2009. PMID: 19702588 Free PMC article. Review.
Cited by
- The validation of the Helicobacter pylori CagA typing by immunohistochemistry: nationwide application in Indonesia.
Miftahussurur M, Doohan D, Syam AF, Nusi IA, Waskito LA, Fauzia KA, Rezkitha YAA, Dewayani A, I'tishom R, Maulahela H, Uchida T, Yamaoka Y. Miftahussurur M, et al. Acta Histochem. 2020 Sep;122(6):151594. doi: 10.1016/j.acthis.2020.151594. Epub 2020 Jul 22. Acta Histochem. 2020. PMID: 32778248 Free PMC article. - Identification of immunodominant antigens from Helicobacter pylori and evaluation of their reactivities with sera from patients with different gastroduodenal pathologies.
Kimmel B, Bosserhoff A, Frank R, Gross R, Goebel W, Beier D. Kimmel B, et al. Infect Immun. 2000 Feb;68(2):915-20. doi: 10.1128/IAI.68.2.915-920.2000. Infect Immun. 2000. PMID: 10639463 Free PMC article. - Helicobacter pylori associated factors in the development of gastric cancer with special reference to the early-onset subtype.
Pucułek M, Machlowska J, Wierzbicki R, Baj J, Maciejewski R, Sitarz R. Pucułek M, et al. Oncotarget. 2018 Jul 24;9(57):31146-31162. doi: 10.18632/oncotarget.25757. eCollection 2018 Jul 24. Oncotarget. 2018. PMID: 30123433 Free PMC article. Review. - Simple method for determination of the number of Helicobacter pylori CagA variable-region EPIYA tyrosine phosphorylation motifs by PCR.
Argent RH, Zhang Y, Atherton JC. Argent RH, et al. J Clin Microbiol. 2005 Feb;43(2):791-5. doi: 10.1128/JCM.43.2.791-795.2005. J Clin Microbiol. 2005. PMID: 15695681 Free PMC article. - Helicobacter pylori cagA gene variants in Malaysians of different ethnicity.
Mohamed R, Hanafiah A, Rose IM, Manaf MR, Abdullah SA, Sagap I, van Belkum A, Yaacob JA. Mohamed R, et al. Eur J Clin Microbiol Infect Dis. 2009 Jul;28(7):865-9. doi: 10.1007/s10096-009-0712-x. Epub 2009 Feb 27. Eur J Clin Microbiol Infect Dis. 2009. PMID: 19247698 Free PMC article.
References
- Blaser M J, Perez-Perez G I, Kleanthous H, Cover T L, Peek R M, Chyou P H, Stemmermann G N, Nomura A. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55:2111–2115. - PubMed
- Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N, Rappuoli R. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;90:5791–5795. - PMC - PubMed
- Dixon M F, Genta R M, Yardley J H, Correa P. Classification and grading of gastritis: the updated Sydney system. Am J Surg Pathol. 1996;20:1161–1181. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases