Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin - PubMed (original) (raw)
Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin
E V Sokurenko et al. Proc Natl Acad Sci U S A. 1998.
Abstract
Conventional wisdom regarding mechanisms of bacterial pathogenesis holds that pathogens arise by external acquisition of distinct virulence factors, whereas determinants shared by pathogens and commensals are considered to be functionally equivalent and have been ignored as genes that could become adapted specifically for virulence. It is shown here, however, that genetic variation in an originally commensal trait, the FimH lectin of type 1 fimbriae, can change the tropism of Escherichia coli, shifting it toward a urovirulent phenotype. Random point mutations in fimH genes that increase binding of the adhesin to mono-mannose residues, structures abundant in the oligosaccharide moieties of urothelial glycoproteins, confer increased virulence in the mouse urinary tract. These mutant FimH variants, however, are characterized by increased sensitivity to soluble inhibitors bathing the oropharyngeal mucosa, the physiological portal of E. coli. This functional trade-off seems to be detrimental for the intestinal ecology of the urovirulent E. coli. Thus, bacterial virulence can be increased by random functional mutations in a commensal trait that are adaptive for a pathologic environment, even at the cost of reduced physiological fitness in the nonpathologic habitat.
Figures
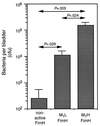
Figure 1
Colonization of mouse bladders by isogenic E. coli expressing nonfunctional FimH, M1L FimH, or M1H FimH subunits. Bars indicate mean cfu per bladder ± SEM. P values indicating level of significance between different groups are indicated.
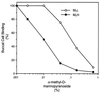
Figure 2
Inhibition of interaction of E. coli and buccal cells by α-methyl-
d
-mannopyranoside.
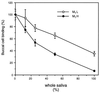
Figure 3
Inhibition of the interaction of E. coli and buccal cells by whole, stimulated human saliva.
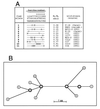
Figure 4
Phylogenetic analysis of FimH alleles. (A) Amino acid sequences of FimH variants. The alleles are listed based on an increasing M1:M3 binding ratio. The residues listed above the 11 alleles are for the amino acids in original FimH sequence (17) that vary in the other fimH alleles. Only polymorphic residues are shown, and the positions are numbered vertically, from −16 to +201. ▵, deleted residues. (B) Inferred phylogenetic network demonstrating evolutionary relationships of the FimH alleles shown in A. Each node represents a distinct FimH allele, numbered as in A. The allele labeled n represents a hypothetical FimH that differs from allele #2 by the substitution of Asp (N) for Tyr (T) in the leader sequence (residue, −16) and phenotypically should be equivalent to allele #2. Internal nodes are shown in bold. The deduced sequences of the 11 FimH proteins exhibit greater than 99% homology, and the network showing their phylogenetic relationships is fully consistent, without any homoplasty. Branch lengths are scaled to the number of amino acids that differ between alleles, as indicated. The deletion of 4 amino acids in FimH allele #10 is considered to be a single event, equivalent to one amino acid substitution.
Similar articles
- Differential stability and trade-off effects of pathoadaptive mutations in the Escherichia coli FimH adhesin.
Weissman SJ, Beskhlebnaya V, Chesnokova V, Chattopadhyay S, Stamm WE, Hooton TM, Sokurenko EV. Weissman SJ, et al. Infect Immun. 2007 Jul;75(7):3548-55. doi: 10.1128/IAI.01963-06. Epub 2007 May 14. Infect Immun. 2007. PMID: 17502398 Free PMC article. - Biofilm formation in a hydrodynamic environment by novel fimh variants and ramifications for virulence.
Schembri MA, Klemm P. Schembri MA, et al. Infect Immun. 2001 Mar;69(3):1322-8. doi: 10.1128/IAI.69.3.1322-1328.2001. Infect Immun. 2001. PMID: 11179294 Free PMC article. - Valency conversion in the type 1 fimbrial adhesin of Escherichia coli.
Sokurenko EV, Schembri MA, Trintchina E, Kjaergaard K, Hasty DL, Klemm P. Sokurenko EV, et al. Mol Microbiol. 2001 Aug;41(3):675-86. doi: 10.1046/j.1365-2958.2001.02545.x. Mol Microbiol. 2001. PMID: 11532135 - Molecular structure of adhesin domains in Escherichia coli fimbriae.
Westerlund-Wikström B, Korhonen TK. Westerlund-Wikström B, et al. Int J Med Microbiol. 2005 Oct;295(6-7):479-86. doi: 10.1016/j.ijmm.2005.06.010. Int J Med Microbiol. 2005. PMID: 16238022 Review. - Enterobacterial adhesins and the case for studying SNPs in bacteria.
Weissman SJ, Moseley SL, Dykhuizen DE, Sokurenko EV. Weissman SJ, et al. Trends Microbiol. 2003 Mar;11(3):115-7. doi: 10.1016/s0966-842x(03)00010-6. Trends Microbiol. 2003. PMID: 12648942 Review.
Cited by
- Uroplakins and their potential applications in urology.
Matuszewski MA, Tupikowski K, Dołowy Ł, Szymańska B, Dembowski J, Zdrojowy R. Matuszewski MA, et al. Cent European J Urol. 2016;69(3):252-257. doi: 10.5173/ceju.2016.638. Epub 2016 Jul 8. Cent European J Urol. 2016. PMID: 27729990 Free PMC article. Review. - Limosilactobacillus fermentum Strain 3872: Antibacterial and Immunoregulatory Properties and Synergy with Prebiotics against Socially Significant Antibiotic-Resistant Infections of Animals and Humans.
Abramov VM, Kosarev IV, Machulin AV, Priputnevich TV, Chikileva IO, Deryusheva EI, Abashina TN, Donetskova AD, Panin AN, Melnikov VG, Suzina NE, Nikonov IN, Selina MV, Khlebnikov VS, Sakulin VK, Vasilenko RN, Samoilenko VA, Uversky VN, Karlyshev AV. Abramov VM, et al. Antibiotics (Basel). 2022 Oct 19;11(10):1437. doi: 10.3390/antibiotics11101437. Antibiotics (Basel). 2022. PMID: 36290095 Free PMC article. - Epidemiologic Investigation of Extra-intestinal pathogenic E. coli (ExPEC) based on PCR phylogenetic group and fimH single nucleotide polymorphisms (SNPs) in China.
Abdallah KS, Cao Y, Wei DJ. Abdallah KS, et al. Int J Mol Epidemiol Genet. 2011;2(4):339-53. Epub 2011 Nov 25. Int J Mol Epidemiol Genet. 2011. PMID: 22199997 Free PMC article. - Temporal colonization and metabolic regulation of the gut microbiome in neonatal oxen at single nucleotide resolution.
Dong Q, Hua D, Wang X, Jiao Y, Liu L, Deng Q, Wu T, Zou H, Zhao C, Wang C, Reng J, Ding L, Hu S, Shi J, Wang Y, Zhang H, Sheng Y, Sun W, Shen Y, Tang L, Kong X, Chen L. Dong Q, et al. ISME J. 2024 Jan 8;18(1):wrad022. doi: 10.1093/ismejo/wrad022. ISME J. 2024. PMID: 38365257 Free PMC article. - Probing genomic diversity and evolution of Escherichia coli O157 by single nucleotide polymorphisms.
Zhang W, Qi W, Albert TJ, Motiwala AS, Alland D, Hyytia-Trees EK, Ribot EM, Fields PI, Whittam TS, Swaminathan B. Zhang W, et al. Genome Res. 2006 Jun;16(6):757-67. doi: 10.1101/gr.4759706. Epub 2006 Apr 10. Genome Res. 2006. PMID: 16606700 Free PMC article.
References
- Falkow S. ASM News. 1997;63:359–365.
- Milkman R. Science. 1973;182:1024–1026. - PubMed
- Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, U.K.: Cambridge Univ. Press; 1983.
- Hartl D L, Dykhuizen D E. In: Population Genetics and Molecular Evolution. Ohta T, Aoki K, editors. Tokyo: Jap. Sci. Soc. Press; 1985. pp. 107–124.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources