Systematic identification of essential genes by in vitro mariner mutagenesis - PubMed (original) (raw)
Systematic identification of essential genes by in vitro mariner mutagenesis
B J Akerley et al. Proc Natl Acad Sci U S A. 1998.
Abstract
Although the complete DNA sequences of several microbial genomes are now available, nearly 40% of the putative genes lack identifiable functions. Comprehensive screens and selections for identifying functional classes of genes are needed to convert sequence data into meaningful biological information. One particularly significant group of bacterial genes consists of those that are essential for growth or viability. Here, we describe a simple system for performing transposon mutagenesis on naturally transformable organisms along with a technique to rapidly identify essential or conditionally essential DNA segments. We show the general utility of this approach by applying it to two human pathogens, Haemophilus influenzae and Streptococcus pneumoniae, in which we detected known essential genes and assigned essentiality to several ORFs of unknown function.
Figures
Figure 1
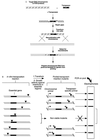
Schematic diagram of the two steps required for GAMBIT (a) Strategy for producing chromosomal mutations by using in vitro transposon mutagenesis. (b) Genetic footprinting for detection of essential genes. Target DNA mutagenized in vitro with the Himar1 transposon was introduced into bacteria by transformation and homologous recombination. Recombinants were selected for drug resistance encoded by the transposon, and insertions in essential genes were lost from the pool during growth. PCR with primers that hybridize to the transposon and to specific chromosomal sites yielded a product corresponding to each mutation in the pool. DNA regions containing no insertions yielded a blank region on electrophoresis gels.
Figure 2
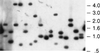
Southern blot analysis of H. influenzae transposon mutants. Genomic DNA was isolated from 16 individual mutants and was digested with _Ase_I, which cleaves once within magellan1. Digested DNA was subjected to agarose gel electrophoresis, was transferred to nitrocellulose, and was hybridized with a probe composed solely of magellan1 minitransposon-derived DNA.
Figure 3
Genetic footprinting of H. influenzae mutant pools. Genetic footprinting was carried out by using a _Himar1_-specific primer and a chromosomal primer. In a, the positions of molecular weight standards are indicated; other panels are labeled with locus names by HI number. In c and d, cells were selected on BXV, MIc, or BXV containing trimethoprim (“Tri”). In f, in vitro mutagenesis of a chromosomal fragment that included the secA gene was performed, and the mutagenized DNA was transformed into both wild-type H. influenzae and an H. influenzae strain containing pSecA.
Figure 4
Essential ORFs of H. influenzae. Five chromosomal segments are shown. ORFs with essential functions are shown in black, ORFs that are nonessential are shown in white, and ORFs in which mutations produce growth attenuation are shown in gray. The direction of transcription for each ORF is shown along with the TIGR designation below the ORF and the closest homologue above the ORF. Conserved hypothetical ORFs of unknown function are designated CH. [*, Essential ORFs that can sustain a highly limited number of discrete insertions (<2/kbp).]
Similar articles
- Identification and analysis of essential genes in Haemophilus influenzae.
Wong SM, Akerley BJ. Wong SM, et al. Methods Mol Biol. 2008;416:27-44. doi: 10.1007/978-1-59745-321-9_3. Methods Mol Biol. 2008. PMID: 18392959 Review. - Genetic footprinting with mariner-based transposition in Pseudomonas aeruginosa.
Wong SM, Mekalanos JJ. Wong SM, et al. Proc Natl Acad Sci U S A. 2000 Aug 29;97(18):10191-6. doi: 10.1073/pnas.97.18.10191. Proc Natl Acad Sci U S A. 2000. PMID: 10963681 Free PMC article. - A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae.
Akerley BJ, Rubin EJ, Novick VL, Amaya K, Judson N, Mekalanos JJ. Akerley BJ, et al. Proc Natl Acad Sci U S A. 2002 Jan 22;99(2):966-71. doi: 10.1073/pnas.012602299. Proc Natl Acad Sci U S A. 2002. PMID: 11805338 Free PMC article. - Genome scanning in Haemophilus influenzae for identification of essential genes.
Reich KA, Chovan L, Hessler P. Reich KA, et al. J Bacteriol. 1999 Aug;181(16):4961-8. doi: 10.1128/JB.181.16.4961-4968.1999. J Bacteriol. 1999. PMID: 10438768 Free PMC article. - Phase variation of lipopolysaccharide in Haemophilus influenzae.
Maskell DJ, Szabo MJ, Butler PD, Williams AE, Moxon ER. Maskell DJ, et al. Res Microbiol. 1991 Jul-Aug;142(6):719-24. doi: 10.1016/0923-2508(91)90086-p. Res Microbiol. 1991. PMID: 1961982 Review.
Cited by
- Bacterial genome engineering using CRISPR-associated transposases.
Gelsinger DR, Vo PLH, Klompe SE, Ronda C, Wang HH, Sternberg SH. Gelsinger DR, et al. Nat Protoc. 2024 Mar;19(3):752-790. doi: 10.1038/s41596-023-00927-3. Epub 2024 Jan 12. Nat Protoc. 2024. PMID: 38216671 Review. - CRISPR-Associated Transposase for Targeted Mutagenesis in Diverse Proteobacteria.
Trujillo Rodríguez L, Ellington AJ, Reisch CR, Chevrette MG. Trujillo Rodríguez L, et al. ACS Synth Biol. 2023 Jul 21;12(7):1989-2003. doi: 10.1021/acssynbio.3c00065. Epub 2023 Jun 27. ACS Synth Biol. 2023. PMID: 37368499 Free PMC article. - High-throughput functional genomics: A (myco)bacterial perspective.
Winkler KR, Mizrahi V, Warner DF, De Wet TJ. Winkler KR, et al. Mol Microbiol. 2023 Aug;120(2):141-158. doi: 10.1111/mmi.15103. Epub 2023 Jun 6. Mol Microbiol. 2023. PMID: 37278255 Free PMC article. Review. - Bacterial genome engineering using CRISPR RNA-guided transposases.
Gelsinger DR, Vo PLH, Klompe SE, Ronda C, Wang H, Sternberg SH. Gelsinger DR, et al. bioRxiv [Preprint]. 2023 Mar 21:2023.03.18.533263. doi: 10.1101/2023.03.18.533263. bioRxiv. 2023. PMID: 36993567 Free PMC article. Updated. Preprint. - Integrated genomics and chemical biology herald an era of sophisticated antibacterial discovery, from defining essential genes to target elucidation.
Warrier T, Romano KP, Clatworthy AE, Hung DT. Warrier T, et al. Cell Chem Biol. 2022 May 19;29(5):716-729. doi: 10.1016/j.chembiol.2022.04.006. Epub 2022 May 5. Cell Chem Biol. 2022. PMID: 35523184 Free PMC article. Review.
References
- Harris S D, Cheng J, Pugh T A, Pringle J R. J Mol Biol. 1992;225:53–65. - PubMed
- Gaiano N, Amsterdam A, Kawakami K, Allende M, Becker T, Hopkins N. Nature (London) 1996;383:829–832. - PubMed
- Murphy C K, Stewart E J, Beckwith J. Gene. 1995;155:1–7. - PubMed
- Blattner F R, Plunkett G R, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. - PubMed
- Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, et al. Nature (London) 1997;388:539–547. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources