E2F3 activity is regulated during the cell cycle and is required for the induction of S phase - PubMed (original) (raw)
E2F3 activity is regulated during the cell cycle and is required for the induction of S phase
G Leone et al. Genes Dev. 1998.
Abstract
Previous work has demonstrated the important role of E2F transcription activity in the induction of S phase during the transition from quiescence to proliferation. In addition to the E2F-dependent activation of a number of genes encoding DNA replication activities such as DNA Pol alpha, we now show that the majority of genes encoding initiation proteins, including Cdc6 and the Mcm proteins, are activated following the stimulation of cell growth and are regulated by E2F. The transcription of a subset of these genes, which includes Cdc6, cyclin E, and cdk2, is also regulated during the cell cycle. Moreover, whereas overall E2F DNA-binding activity accumulates during the initial G1 following a growth stimulus, only E2F3-binding activity reaccumulates at subsequent G1/S transitions, coincident with the expression of the cell-cycle-regulated subset of E2F-target genes. Finally, we show that immunodepletion of E2F3 activity inhibits the induction of S phase in proliferating cells. We propose that E2F3 activity plays an important role during the cell cycle of proliferating cells, controlling the expression of genes whose products are rate limiting for initiation of DNA replication, thereby imparting a more dramatic control of entry into S phase than would otherwise be achieved by post-transcriptional control alone.
Figures
Figure 1
Growth regulated expression of genes encoding DNA replication proteins. (A) Quiescent Ref 52 cells (Q) were stimulated with media containing 10% serum. Cells were harvested at the indicated times, processed for Northern analysis as described in Materials and Methods, and hybridized with the indicated probes. The position of the cell with respect to cell cycle is indicated below based on FACS analysis of similarly treated samples. (B) Quiescent cells were infected with recombinant adenoviruses expressing either E2F1, E2F2, or E2F3 proteins, or with a control virus containing an empty expression cassette (m.o.i. of 100, 100, 200, and 100, respectively). Cells were harvested 18 hr postinfection and processed and analyzed as in A. A portion of the cells that were infected with the control virus were stimulated with media containing 10% serum for an additional 18 hr (Con +).
Figure 1
Growth regulated expression of genes encoding DNA replication proteins. (A) Quiescent Ref 52 cells (Q) were stimulated with media containing 10% serum. Cells were harvested at the indicated times, processed for Northern analysis as described in Materials and Methods, and hybridized with the indicated probes. The position of the cell with respect to cell cycle is indicated below based on FACS analysis of similarly treated samples. (B) Quiescent cells were infected with recombinant adenoviruses expressing either E2F1, E2F2, or E2F3 proteins, or with a control virus containing an empty expression cassette (m.o.i. of 100, 100, 200, and 100, respectively). Cells were harvested 18 hr postinfection and processed and analyzed as in A. A portion of the cells that were infected with the control virus were stimulated with media containing 10% serum for an additional 18 hr (Con +).
Figure 2
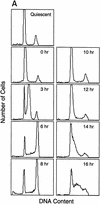
A subset of E2F targets, including Cdc6, Cyclin E, and Cdk2, are regulated during the cell cycle. (A) Quiescent REF 52 cells were stimulated with media containing 10% serum in the presence of 2 m
m
HU. After 21 hr, the G1/S-arrested cells were washed free of HU and then grown in medium containing 10% serum. At the indicated times following the release from the HU block, cells were harvested, stained with propidium iodide, and processed for flow cytometry as described in Materials and Methods. (B) Quiescent REF52 cells (Q) or samples harvested at the indicated times following the release from the HU block were processed for Northern analysis as described in Materials and Methods. Northern blots were hybridized with the indicated probes. Notably, two E2F3-specific RNAs are consistently detected by Northern analysis; the slower-migrating species represents the growth-regulated RNA that coincides with the accumulation of E2F3 protein (see Fig. 5A; data not shown). (C) HeLa cells were arrested at G1/S by incubation with 2 m
m
thymidine for 16 hr, washed, and released in media containing 10% fetal calf serum. At the indicated times, cells were harvested and prepared for FACS analysis as described in Materials and Methods. (D) At the indicated times following release from the thymidine block, HeLa cells were harvested and processed for Northern analysis. The blot was probed with a human specific Cdc6 probe. Blots were stained with methylene blue to confirm equal loading. (A) Asynchronous cells. (E) At the indicated times following the release of HeLa cells from a thymidine block, nuclei were isolated and used for nuclear run-on transcription assays as described in Materials and Methods. The Cdc6-specific transcription rate is presented relative to the transcription rate of the β-actin control. Data are presented as a mean of six independent determinations.
Figure 2
A subset of E2F targets, including Cdc6, Cyclin E, and Cdk2, are regulated during the cell cycle. (A) Quiescent REF 52 cells were stimulated with media containing 10% serum in the presence of 2 m
m
HU. After 21 hr, the G1/S-arrested cells were washed free of HU and then grown in medium containing 10% serum. At the indicated times following the release from the HU block, cells were harvested, stained with propidium iodide, and processed for flow cytometry as described in Materials and Methods. (B) Quiescent REF52 cells (Q) or samples harvested at the indicated times following the release from the HU block were processed for Northern analysis as described in Materials and Methods. Northern blots were hybridized with the indicated probes. Notably, two E2F3-specific RNAs are consistently detected by Northern analysis; the slower-migrating species represents the growth-regulated RNA that coincides with the accumulation of E2F3 protein (see Fig. 5A; data not shown). (C) HeLa cells were arrested at G1/S by incubation with 2 m
m
thymidine for 16 hr, washed, and released in media containing 10% fetal calf serum. At the indicated times, cells were harvested and prepared for FACS analysis as described in Materials and Methods. (D) At the indicated times following release from the thymidine block, HeLa cells were harvested and processed for Northern analysis. The blot was probed with a human specific Cdc6 probe. Blots were stained with methylene blue to confirm equal loading. (A) Asynchronous cells. (E) At the indicated times following the release of HeLa cells from a thymidine block, nuclei were isolated and used for nuclear run-on transcription assays as described in Materials and Methods. The Cdc6-specific transcription rate is presented relative to the transcription rate of the β-actin control. Data are presented as a mean of six independent determinations.
Figure 2
A subset of E2F targets, including Cdc6, Cyclin E, and Cdk2, are regulated during the cell cycle. (A) Quiescent REF 52 cells were stimulated with media containing 10% serum in the presence of 2 m
m
HU. After 21 hr, the G1/S-arrested cells were washed free of HU and then grown in medium containing 10% serum. At the indicated times following the release from the HU block, cells were harvested, stained with propidium iodide, and processed for flow cytometry as described in Materials and Methods. (B) Quiescent REF52 cells (Q) or samples harvested at the indicated times following the release from the HU block were processed for Northern analysis as described in Materials and Methods. Northern blots were hybridized with the indicated probes. Notably, two E2F3-specific RNAs are consistently detected by Northern analysis; the slower-migrating species represents the growth-regulated RNA that coincides with the accumulation of E2F3 protein (see Fig. 5A; data not shown). (C) HeLa cells were arrested at G1/S by incubation with 2 m
m
thymidine for 16 hr, washed, and released in media containing 10% fetal calf serum. At the indicated times, cells were harvested and prepared for FACS analysis as described in Materials and Methods. (D) At the indicated times following release from the thymidine block, HeLa cells were harvested and processed for Northern analysis. The blot was probed with a human specific Cdc6 probe. Blots were stained with methylene blue to confirm equal loading. (A) Asynchronous cells. (E) At the indicated times following the release of HeLa cells from a thymidine block, nuclei were isolated and used for nuclear run-on transcription assays as described in Materials and Methods. The Cdc6-specific transcription rate is presented relative to the transcription rate of the β-actin control. Data are presented as a mean of six independent determinations.
Figure 2
A subset of E2F targets, including Cdc6, Cyclin E, and Cdk2, are regulated during the cell cycle. (A) Quiescent REF 52 cells were stimulated with media containing 10% serum in the presence of 2 m
m
HU. After 21 hr, the G1/S-arrested cells were washed free of HU and then grown in medium containing 10% serum. At the indicated times following the release from the HU block, cells were harvested, stained with propidium iodide, and processed for flow cytometry as described in Materials and Methods. (B) Quiescent REF52 cells (Q) or samples harvested at the indicated times following the release from the HU block were processed for Northern analysis as described in Materials and Methods. Northern blots were hybridized with the indicated probes. Notably, two E2F3-specific RNAs are consistently detected by Northern analysis; the slower-migrating species represents the growth-regulated RNA that coincides with the accumulation of E2F3 protein (see Fig. 5A; data not shown). (C) HeLa cells were arrested at G1/S by incubation with 2 m
m
thymidine for 16 hr, washed, and released in media containing 10% fetal calf serum. At the indicated times, cells were harvested and prepared for FACS analysis as described in Materials and Methods. (D) At the indicated times following release from the thymidine block, HeLa cells were harvested and processed for Northern analysis. The blot was probed with a human specific Cdc6 probe. Blots were stained with methylene blue to confirm equal loading. (A) Asynchronous cells. (E) At the indicated times following the release of HeLa cells from a thymidine block, nuclei were isolated and used for nuclear run-on transcription assays as described in Materials and Methods. The Cdc6-specific transcription rate is presented relative to the transcription rate of the β-actin control. Data are presented as a mean of six independent determinations.
Figure 2
A subset of E2F targets, including Cdc6, Cyclin E, and Cdk2, are regulated during the cell cycle. (A) Quiescent REF 52 cells were stimulated with media containing 10% serum in the presence of 2 m
m
HU. After 21 hr, the G1/S-arrested cells were washed free of HU and then grown in medium containing 10% serum. At the indicated times following the release from the HU block, cells were harvested, stained with propidium iodide, and processed for flow cytometry as described in Materials and Methods. (B) Quiescent REF52 cells (Q) or samples harvested at the indicated times following the release from the HU block were processed for Northern analysis as described in Materials and Methods. Northern blots were hybridized with the indicated probes. Notably, two E2F3-specific RNAs are consistently detected by Northern analysis; the slower-migrating species represents the growth-regulated RNA that coincides with the accumulation of E2F3 protein (see Fig. 5A; data not shown). (C) HeLa cells were arrested at G1/S by incubation with 2 m
m
thymidine for 16 hr, washed, and released in media containing 10% fetal calf serum. At the indicated times, cells were harvested and prepared for FACS analysis as described in Materials and Methods. (D) At the indicated times following release from the thymidine block, HeLa cells were harvested and processed for Northern analysis. The blot was probed with a human specific Cdc6 probe. Blots were stained with methylene blue to confirm equal loading. (A) Asynchronous cells. (E) At the indicated times following the release of HeLa cells from a thymidine block, nuclei were isolated and used for nuclear run-on transcription assays as described in Materials and Methods. The Cdc6-specific transcription rate is presented relative to the transcription rate of the β-actin control. Data are presented as a mean of six independent determinations.
Figure 3
Cell cycle regulation of Cyclin E, Cdk2, and Cdc6 protein accumulation. (A) Protein lysates (10 μg per lane) derived from REF52 cells treated as in the HU arrest/release experiments decribed in Fig. 2A were subjected to 10% SDS-PAGE, Western blotted, and probed with either cyclin E- or cdk2-specific antibodies as indicated. (B) Protein samples (60 μg per lane) from HeLa cells treated as in the thymidine arrest/release experiment described in Fig. 2C were subjected to 10% SDS-PAGE, transferred to nitrocellulose, and probed with antibodies specific to human Cdc6 protein.
Figure 3
Cell cycle regulation of Cyclin E, Cdk2, and Cdc6 protein accumulation. (A) Protein lysates (10 μg per lane) derived from REF52 cells treated as in the HU arrest/release experiments decribed in Fig. 2A were subjected to 10% SDS-PAGE, Western blotted, and probed with either cyclin E- or cdk2-specific antibodies as indicated. (B) Protein samples (60 μg per lane) from HeLa cells treated as in the thymidine arrest/release experiment described in Fig. 2C were subjected to 10% SDS-PAGE, transferred to nitrocellulose, and probed with antibodies specific to human Cdc6 protein.
Figure 4
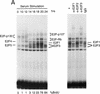
Cell cycle control of E2F activity. (A) Nuclear extracts prepared at various times following the stimulation of quiescent REF52 cells (Q) were assayed for E2F DNA-binding activity by electrophoretic mobility-shift assays (EMSA) using an E2F-specific 32P-labeled DNA probe (left). Cells similarly stimulated were incubated with BrdU (10 μ
m
), fixed at the indicated times, subsequently immunostained with BrdU-specific antibodies, and visualized by immunofluorescent microscopy. The percentage of BrdU-positive cells at each time point is indicated below the DNA-shift gel. The nuclear extract sample from the 20-hr time point presented at left (G1/S sample) was incubated with either IgG-, E2F1-, E2F2-, or E2F3-specific antibodies prior to being subjected similarly to EMSA (right). The E2F4- and E2F5-specific bands indicated on the DNA gel-shift have been identified similarly using specific antibodies against the respective proteins (data not shown). We have been unable to identify an E2F2-specific DNA-binding activity in these assays. (B) Nuclear extracts prepared at various times following the release of cells from an HU block as described in Fig. 2A, as well as from quiescent (Q) REF52 cells, were assayed for E2F DNA-binding activity by EMSA using an E2F-specific 32P-labeled DNA probe (left). The position of the cells with respect to cell cycle is indicated below based on the FACS analysis shown in Fig. 2A. Cytoplasmic extracts prepared from the same time point samples were assayed for E2F-binding activity by EMSA using the same E2F-specific probe (right). (C) Ref52 cells were brought to quiescence by serum starvation and then stimulated to grow by addition of fresh medium with serum. Samples were taken at the indicated times and processed for FACS analysis as described in Materials and Methods. (D) Nuclear extracts prepared at various times following serum stimulation and assayed for E2F DNA-binding activity by EMSA using an E2F-specific 32P-labeled DNA probe.
Figure 4
Cell cycle control of E2F activity. (A) Nuclear extracts prepared at various times following the stimulation of quiescent REF52 cells (Q) were assayed for E2F DNA-binding activity by electrophoretic mobility-shift assays (EMSA) using an E2F-specific 32P-labeled DNA probe (left). Cells similarly stimulated were incubated with BrdU (10 μ
m
), fixed at the indicated times, subsequently immunostained with BrdU-specific antibodies, and visualized by immunofluorescent microscopy. The percentage of BrdU-positive cells at each time point is indicated below the DNA-shift gel. The nuclear extract sample from the 20-hr time point presented at left (G1/S sample) was incubated with either IgG-, E2F1-, E2F2-, or E2F3-specific antibodies prior to being subjected similarly to EMSA (right). The E2F4- and E2F5-specific bands indicated on the DNA gel-shift have been identified similarly using specific antibodies against the respective proteins (data not shown). We have been unable to identify an E2F2-specific DNA-binding activity in these assays. (B) Nuclear extracts prepared at various times following the release of cells from an HU block as described in Fig. 2A, as well as from quiescent (Q) REF52 cells, were assayed for E2F DNA-binding activity by EMSA using an E2F-specific 32P-labeled DNA probe (left). The position of the cells with respect to cell cycle is indicated below based on the FACS analysis shown in Fig. 2A. Cytoplasmic extracts prepared from the same time point samples were assayed for E2F-binding activity by EMSA using the same E2F-specific probe (right). (C) Ref52 cells were brought to quiescence by serum starvation and then stimulated to grow by addition of fresh medium with serum. Samples were taken at the indicated times and processed for FACS analysis as described in Materials and Methods. (D) Nuclear extracts prepared at various times following serum stimulation and assayed for E2F DNA-binding activity by EMSA using an E2F-specific 32P-labeled DNA probe.
Figure 4
Cell cycle control of E2F activity. (A) Nuclear extracts prepared at various times following the stimulation of quiescent REF52 cells (Q) were assayed for E2F DNA-binding activity by electrophoretic mobility-shift assays (EMSA) using an E2F-specific 32P-labeled DNA probe (left). Cells similarly stimulated were incubated with BrdU (10 μ
m
), fixed at the indicated times, subsequently immunostained with BrdU-specific antibodies, and visualized by immunofluorescent microscopy. The percentage of BrdU-positive cells at each time point is indicated below the DNA-shift gel. The nuclear extract sample from the 20-hr time point presented at left (G1/S sample) was incubated with either IgG-, E2F1-, E2F2-, or E2F3-specific antibodies prior to being subjected similarly to EMSA (right). The E2F4- and E2F5-specific bands indicated on the DNA gel-shift have been identified similarly using specific antibodies against the respective proteins (data not shown). We have been unable to identify an E2F2-specific DNA-binding activity in these assays. (B) Nuclear extracts prepared at various times following the release of cells from an HU block as described in Fig. 2A, as well as from quiescent (Q) REF52 cells, were assayed for E2F DNA-binding activity by EMSA using an E2F-specific 32P-labeled DNA probe (left). The position of the cells with respect to cell cycle is indicated below based on the FACS analysis shown in Fig. 2A. Cytoplasmic extracts prepared from the same time point samples were assayed for E2F-binding activity by EMSA using the same E2F-specific probe (right). (C) Ref52 cells were brought to quiescence by serum starvation and then stimulated to grow by addition of fresh medium with serum. Samples were taken at the indicated times and processed for FACS analysis as described in Materials and Methods. (D) Nuclear extracts prepared at various times following serum stimulation and assayed for E2F DNA-binding activity by EMSA using an E2F-specific 32P-labeled DNA probe.
Figure 4
Cell cycle control of E2F activity. (A) Nuclear extracts prepared at various times following the stimulation of quiescent REF52 cells (Q) were assayed for E2F DNA-binding activity by electrophoretic mobility-shift assays (EMSA) using an E2F-specific 32P-labeled DNA probe (left). Cells similarly stimulated were incubated with BrdU (10 μ
m
), fixed at the indicated times, subsequently immunostained with BrdU-specific antibodies, and visualized by immunofluorescent microscopy. The percentage of BrdU-positive cells at each time point is indicated below the DNA-shift gel. The nuclear extract sample from the 20-hr time point presented at left (G1/S sample) was incubated with either IgG-, E2F1-, E2F2-, or E2F3-specific antibodies prior to being subjected similarly to EMSA (right). The E2F4- and E2F5-specific bands indicated on the DNA gel-shift have been identified similarly using specific antibodies against the respective proteins (data not shown). We have been unable to identify an E2F2-specific DNA-binding activity in these assays. (B) Nuclear extracts prepared at various times following the release of cells from an HU block as described in Fig. 2A, as well as from quiescent (Q) REF52 cells, were assayed for E2F DNA-binding activity by EMSA using an E2F-specific 32P-labeled DNA probe (left). The position of the cells with respect to cell cycle is indicated below based on the FACS analysis shown in Fig. 2A. Cytoplasmic extracts prepared from the same time point samples were assayed for E2F-binding activity by EMSA using the same E2F-specific probe (right). (C) Ref52 cells were brought to quiescence by serum starvation and then stimulated to grow by addition of fresh medium with serum. Samples were taken at the indicated times and processed for FACS analysis as described in Materials and Methods. (D) Nuclear extracts prepared at various times following serum stimulation and assayed for E2F DNA-binding activity by EMSA using an E2F-specific 32P-labeled DNA probe.
Figure 5
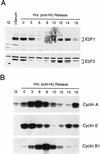
Cell cycle control of E2F protein accumulation. (A) Quiescent REF52 cells (Q), or REF52 cells synchronized at G1/S by HU treatment as described in Fig. 2A, were harvested at the indicated times following the release of the G1/S block, and processed for Western blot analysis using either E2F1- or E2F3-specific antibodies. For comparison, samples from quiescent cells (Q) or quiescent cells stimulated for 19 hr with 10% serum (+serum) were also included. (B) Aliquots from the same HU-treated samples, or from quiescent cells (Q) were processed for cyclin A-, cyclin E-, or cyclin B1-dependent kinase activity by immunoprecipitation of kinase complexes using the indicated antibodies specific for the respective cyclin activities and using histone H1 as a substrate. The kinase reactions were analyzed by 10% SDS-PAGE.
Figure 6
Inhibition of E2F3 activity inhibits the cell cycle induction of S phase. (A) Quiescent REF52 cells were infected with either Ad–E2F1, Ad–E2F3, or Ad–Con (m.o.i. of 100, 200, and 100, respectively). Cells were microinjected 4 hr postinfection with IgG, or with E2F1- or E2F3-specific antibodies (at an antibody concentration of 1 μg/ml, containing fluorescein-conjugated dextran as a marker for detecting injected cells). BrdU was added 12 hr later (10 μ
m
) and cells were incubated an additional 4 hr prior to fixation and immunostaining with BrdU-specific antibodies. Microinjected cells were visualized by fluorescent microscopy and the percentage of cells staining positively for BrdU is presented above. Approximately 150–250 cells were microinjected in two separate experiments, a representative experiment is shown. (−) The quantitation of uninjected cells in the same tissue-culture plate. (B) REF52 cells were synchronized by HU treatment as described in Fig. 2A, and 5–7 hr following the release from the G1/S block, a time when cells are predominantly in G2, cells were microinjected with IgG, or either with E2F1- or E2F3-specific antibodies. BrdU was then added to the cells 14 hr after the HU release and incubated for a further 3 hr, after which cells were fixed, immunostained, and quantitated for BrdU incorporation as in A. Approximately 150–250 cells were microinjected for each condition, and a representative experiment (similar results were obtained in five independent experiments) is presented. (−) The quantitation of uninjected cells in the same tissue culture plate.
Similar articles
- Cdk2-dependent and -independent pathways in E2F-mediated S phase induction.
Arata Y, Fujita M, Ohtani K, Kijima S, Kato JY. Arata Y, et al. J Biol Chem. 2000 Mar 3;275(9):6337-45. doi: 10.1074/jbc.275.9.6337. J Biol Chem. 2000. PMID: 10692433 - Collaborative role of E2F transcriptional activity and G1 cyclindependent kinase activity in the induction of S phase.
Leone G, DeGregori J, Jakoi L, Cook JG, Nevins JR. Leone G, et al. Proc Natl Acad Sci U S A. 1999 Jun 8;96(12):6626-31. doi: 10.1073/pnas.96.12.6626. Proc Natl Acad Sci U S A. 1999. PMID: 10359762 Free PMC article. - Regulation of E2F transcription by cyclin E-Cdk2 kinase mediated through p300/CBP co-activators.
Morris L, Allen KE, La Thangue NB. Morris L, et al. Nat Cell Biol. 2000 Apr;2(4):232-9. doi: 10.1038/35008660. Nat Cell Biol. 2000. PMID: 10783242 - Cell cycle and transcriptional control of human myeloid leukemic cells by transforming growth factor beta.
Hu X, Zuckerman KS. Hu X, et al. Leuk Lymphoma. 2000 Jul;38(3-4):235-46. doi: 10.3109/10428190009087015. Leuk Lymphoma. 2000. PMID: 10830731 Review. - Induction of S phase by G1 regulatory factors.
Kato J. Kato J. Front Biosci. 1999 Dec 1;4:D787-92. doi: 10.2741/kato. Front Biosci. 1999. PMID: 10577389 Review.
Cited by
- E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells.
Chong JL, Wenzel PL, Sáenz-Robles MT, Nair V, Ferrey A, Hagan JP, Gomez YM, Sharma N, Chen HZ, Ouseph M, Wang SH, Trikha P, Culp B, Mezache L, Winton DJ, Sansom OJ, Chen D, Bremner R, Cantalupo PG, Robinson ML, Pipas JM, Leone G. Chong JL, et al. Nature. 2009 Dec 17;462(7275):930-4. doi: 10.1038/nature08677. Nature. 2009. PMID: 20016602 Free PMC article. - Activation of Cdc6 by MyoD is associated with the expansion of quiescent myogenic satellite cells.
Zhang K, Sha J, Harter ML. Zhang K, et al. J Cell Biol. 2010 Jan 11;188(1):39-48. doi: 10.1083/jcb.200904144. Epub 2010 Jan 4. J Cell Biol. 2010. PMID: 20048262 Free PMC article. - Distinct roles of E2F proteins in vascular smooth muscle cell proliferation and intimal hyperplasia.
Giangrande PH, Zhang J, Tanner A, Eckhart AD, Rempel RE, Andrechek ER, Layzer JM, Keys JR, Hagen PO, Nevins JR, Koch WJ, Sullenger BA. Giangrande PH, et al. Proc Natl Acad Sci U S A. 2007 Aug 7;104(32):12988-93. doi: 10.1073/pnas.0704754104. Epub 2007 Jul 25. Proc Natl Acad Sci U S A. 2007. PMID: 17652516 Free PMC article. - E2f1, E2f2, and E2f3 control E2F target expression and cellular proliferation via a p53-dependent negative feedback loop.
Timmers C, Sharma N, Opavsky R, Maiti B, Wu L, Wu J, Orringer D, Trikha P, Saavedra HI, Leone G. Timmers C, et al. Mol Cell Biol. 2007 Jan;27(1):65-78. doi: 10.1128/MCB.02147-05. Mol Cell Biol. 2007. PMID: 17167174 Free PMC article. - Serum response factor plays an important role in the mechanically overloaded plantaris muscle of rats.
Sakuma K, Nishikawa J, Nakao R, Nakano H, Sano M, Yasuhara M. Sakuma K, et al. Histochem Cell Biol. 2003 Feb;119(2):149-60. doi: 10.1007/s00418-003-0499-2. Epub 2003 Feb 6. Histochem Cell Biol. 2003. PMID: 12610734
References
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. - PubMed
- DeGregori J, Leone G, Ohtani K, Miron A, Nevins JR. E2F1 accumulation bypasses a G1 arrest resulting from the inhibition of G1 cyclin-dependent kinase activity. Genes & Dev. 1995b;9:2873–2887. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources