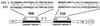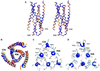Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides - PubMed (original) (raw)
Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides
V N Malashkevich et al. Proc Natl Acad Sci U S A. 1998.
Abstract
The gp41 subunit of the envelope protein complex from human and simian immunodeficiency viruses (HIV and SIV) mediates membrane fusion during viral entry. The crystal structure of the HIV-1 gp41 ectodomain core in its proposed fusion-active state is a six-helix bundle. Here we have reconstituted the core of the SIV gp41 ectodomain with two synthetic peptides called SIV N36 and SIV C34, which form a highly helical trimer of heterodimers. The 2.2 A resolution crystal structure of this SIV N36/C34 complex is very similar to the analogous structure in HIV-1 gp41. In both structures, three N36 helices form a central trimeric coiled coil. Three C34 helices pack in an antiparallel orientation into highly conserved, hydrophobic grooves along the surface of this coiled coil. The conserved nature of the N36-C34 interface suggests that the HIV-1 and SIV peptides are functionally interchangeable. Indeed, a heterotypic complex between HIV-1 N36 and SIV C34 peptides is highly helical and stable. Moreover, as with HIV-1 C34, the SIV C34 peptide is a potent inhibitor of HIV-1 infection. These results identify conserved packing interactions between the N and C helices of gp41 and have implications for the development of C peptide analogs with broad inhibitory activity.
Figures
Figure 1
A schematic view of gp41, showing the locations of the N36 and C34 peptides from SIV and HIV-1. Identical residues between the SIV and HIV-1 peptides are indicated with a bar, and similar residues are indicated by dots. To facilitate comparisons between the HIV-1 and SIV structures, the peptide residues are numbered according to their positions in HIV-1 gp160 (HXB2 strain; GenBank accession no. K03455). For the SIV peptides (SIV Mac239 strain; GenBank accession no. M33262), N36 actually corresponds to positions 559–594, and C34 corresponds to 637–670. The 4,3 hydrophobic repeats are indicated with black shading. FP, fusion peptide; S—S, disulfide bond; TM, transmembrane region; CYTO, cytoplasmic domain.
Figure 2
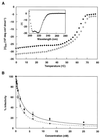
CD spectra of SIV N36 and C34 peptides. CD spectra of SIV N36 (▿), SIV C34 (▵), and the SIV N36 C34 complex (♦) in PBS. The predicted spectrum for noninteracting peptides (⋄) is shown for comparison. Inset shows the thermal dependence of the CD signal at 222 nm for the SIV N36/C34 complex.
Figure 3
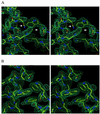
Stereoviews of representative electron density maps. (A) Initial 2Fo-Fc map after density modification and phase improvement with
dm
(19) superimposed onto the initial molecular replacement solution model. A shift of the N36 helix (left side of figure) and other clear differences between the trial model and the observed density are seen (white arrows). (B) Final 2Fo-Fc map superimposed onto the refined model. Water molecules are indicated with small, red balls. Both maps are contoured at 1.5 σ. Figures were generated with
o
(20).
Figure 4
Structure of SIV N36/C34 complex and comparison with that of HIV-1. (A) Stereoview from the side. The amino termini of N36 and the carboxyl termini of C34 are at the top of the figure. Helices in the N36 coiled-coil trimer from SIV (blue) and HIV-1 (yellow) were used for the superposition. The inner N36 coiled-coil trimers superimpose well; however, a pronounced shift in the central region of the outer C34 helices is visible. (B) View from the top, along the noncrystallographic 3-fold axis. The same color coding is used. (C) A cross-section of the SIV ectodomain composed almost exclusively of glutamine residues, six of which are conserved in the HIV ectodomain sequence and two that are conservatively replaced by asparagines. Glutamine side chains are involved in a web of hydrogen bonds that also involve some carbonyl oxygens of the main chain. (D) A cross-section of the SIV N36/C34 structure near the conserved, deep hydrophobic cavity involving three tryptophan residues. Figures were generated with
grasp
(28).
Figure 5
The conserved hydrophobic cavity on the surface of the N36 coiled coil. (A) Structural overlay of the published HIV-1 gp41 ectodomain structures. The three structures [Chan et al. (5), yellow; Weissenhorn et al. (6), red; and Tan et al. (7), green] deviate substantially in this region. (B) Interactions in the N36 hydrophobic cavity of SIV (blue) are most similar to the Chan et al. (5) HIV-1 structure (yellow). The same superposition as in Fig. 4 A and B was used. The SIV N36 coiled coil is represented as a molecular surface, and the C34 helices are shown as ribbons with selected side chains. The molecular surface is colored white for residues that are identical in SIV and HIV, and green for residues that are not identical. Figures were generated with
grasp
(28).
Figure 6
HIV/SIV heterotypic complexes. (A) Thermal dependence of the CD signal at 222 nm for the HIV N36/SIV C34 heterotypic complex (♦) and the HIV N36/HIV C34 complex (○). Inset shows the CD spectrum for the HIV N36/SIV C34 heterotypic complex. (B) Inhibition of HIV-1 (HXB2 strain) infection by SIV C34 (♦ and solid line) and HIV C34 peptide (○ and dashed line). Error bars indicate the SE from triplicate experiments.
Similar articles
- Buried polar interactions and conformational stability in the simian immunodeficiency virus (SIV) gp41 core.
Ji H, Bracken C, Lu M. Ji H, et al. Biochemistry. 2000 Feb 1;39(4):676-85. doi: 10.1021/bi991893e. Biochemistry. 2000. PMID: 10651632 - The stability of the intact envelope glycoproteins is a major determinant of sensitivity of HIV/SIV to peptidic fusion inhibitors.
Gallo SA, Sackett K, Rawat SS, Shai Y, Blumenthal R. Gallo SA, et al. J Mol Biol. 2004 Jun 25;340(1):9-14. doi: 10.1016/j.jmb.2004.04.027. J Mol Biol. 2004. PMID: 15184018 - Multimerized HIV-gp41-derived peptides as fusion inhibitors and vaccines.
Nomura W, Mizuguchi T, Tamamura H. Nomura W, et al. Biopolymers. 2016 Nov 4;106(4):622-8. doi: 10.1002/bip.22782. Biopolymers. 2016. PMID: 26583370 Review. - N- and C-domains of HIV-1 gp41: mutation, structure and functions.
Dong XN, Xiao Y, Dierich MP, Chen YH. Dong XN, et al. Immunol Lett. 2001 Jan 15;75(3):215-20. doi: 10.1016/s0165-2478(00)00302-3. Immunol Lett. 2001. PMID: 11166378 Review.
Cited by
- Biomedical applications of glycosylphosphatidylinositol-anchored proteins.
Heider S, Dangerfield JA, Metzner C. Heider S, et al. J Lipid Res. 2016 Oct;57(10):1778-1788. doi: 10.1194/jlr.R070201. Epub 2016 Aug 19. J Lipid Res. 2016. PMID: 27542385 Free PMC article. Review. - Structural characterization of the human respiratory syncytial virus fusion protein core.
Zhao X, Singh M, Malashkevich VN, Kim PS. Zhao X, et al. Proc Natl Acad Sci U S A. 2000 Dec 19;97(26):14172-7. doi: 10.1073/pnas.260499197. Proc Natl Acad Sci U S A. 2000. PMID: 11106388 Free PMC article. - Antiviral activity and conformational features of an octapeptide derived from the membrane-proximal ectodomain of the feline immunodeficiency virus transmembrane glycoprotein.
Giannecchini S, Di Fenza A, D'Ursi AM, Matteucci D, Rovero P, Bendinelli M. Giannecchini S, et al. J Virol. 2003 Mar;77(6):3724-33. doi: 10.1128/jvi.77.6.3724-3733.2003. J Virol. 2003. PMID: 12610147 Free PMC article. - Conserved salt bridge between the N- and C-terminal heptad repeat regions of the human immunodeficiency virus type 1 gp41 core structure is critical for virus entry and inhibition.
He Y, Liu S, Li J, Lu H, Qi Z, Liu Z, Debnath AK, Jiang S. He Y, et al. J Virol. 2008 Nov;82(22):11129-39. doi: 10.1128/JVI.01060-08. Epub 2008 Sep 3. J Virol. 2008. PMID: 18768964 Free PMC article. - HIV-1 Entry, Inhibitors, and Resistance.
Lobritz MA, Ratcliff AN, Arts EJ. Lobritz MA, et al. Viruses. 2010 May;2(5):1069-1105. doi: 10.3390/v2051069. Epub 2010 Apr 29. Viruses. 2010. PMID: 21994672 Free PMC article.
References
- Chan D C, Kim P S. Cell. 1998;93:681–684. - PubMed
- Lu M, Blacklow S C, Kim P S. Nat Struct Biol. 1995;2:1075–1082. - PubMed
- Lu M, Kim P S. J Biomol Struct Dyn. 1997;15:465–471. - PubMed
- Blacklow S C, Lu M, Kim P S. Biochemistry. 1995;34:14955–14962. - PubMed
- Chan D C, Fass D, Berger J M, Kim P S. Cell. 1997;89:263–273. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources