Evolutionary parameters of the transcribed mammalian genome: an analysis of 2,820 orthologous rodent and human sequences - PubMed (original) (raw)
Evolutionary parameters of the transcribed mammalian genome: an analysis of 2,820 orthologous rodent and human sequences
W Makalowski et al. Proc Natl Acad Sci U S A. 1998.
Abstract
We have rigorously defined 2,820 orthologous mRNA and protein sequence pairs from rats, mice, and humans. Evolutionary rate analyses indicate that mammalian genes are evolving 17-30% more slowly than previous textbook values. Data are presented on the average properties of mRNA and protein sequences, on variations in sequence conservation in coding and noncoding regions, and on the absolute and relative frequencies of repetitive elements and splice sites in untranslated regions of mRNAs. Our data set contains 1,880 unique human/rodent sequence pairs that represent about 2-4% of all mammalian genes. Of the 1,880 human orthologs, 70% are present on a new gene map of the human genome, thus providing a valuable resource for cross-referencing human and rodent genomes. In addition to comparative mapping, these results have practical applications in the interpretation of noncoding sequence conservation between syntenic regions of human and mouse genomic sequence, and in the design and calibration of gene expression arrays.
Figures
Figure 1
Data sets of orthologous sequence pairs.
Figure 2
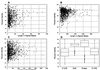
Distributions of lengths and degrees of sequence conservation for 1,212 aligned orthologous rat and human mRNA and protein sequences. (A–C) Scatter plots of results for 5′ UTRs (A), CDSs (B), and 3′ UTRs (C). (D) Box plots of sequence conservation by region for aligned rat and human mRNAs and encoded proteins. For each category, the central box depicts the middle 50% of the data between the 25th and 75th percentile, and the enclosed horizontal line represents the median value of the distribution. Extreme values are indicated by circles that occur outside the main bodies of data.
Figure 3
Distributions of lengths and degrees of sequence conservation for 470 aligned orthologous mouse and rat mRNA and protein sequences. (A–C) Scatter plots of results for 5′ UTRs (A), CDSs (B), and 3′ UTRs (C). (D) Box plots as described in the legend to Fig. 2.
Figure 4
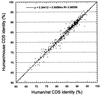
Correlation of coding sequence identities between orthologous human/mouse and human/rat sequence pairs.
Figure 5
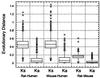
Analysis of evolutionary distances for orthologous sequence pairs.
Figure 6
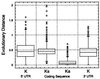
Analysis of evolutionary distances in untranslated and coding regions of human–rodent mRNA sequences.
Similar articles
- Evolutionary dynamics of mammalian mRNA untranslated regions by comparative analysis of orthologous human, artiodactyl and rodent gene pairs.
Larizza A, Makalowski W, Pesole G, Saccone C. Larizza A, et al. Comput Chem. 2002 Jul;26(5):479-90. doi: 10.1016/s0097-8485(02)00009-8. Comput Chem. 2002. PMID: 12144177 - Comparative analysis of 1196 orthologous mouse and human full-length mRNA and protein sequences.
Makałowski W, Zhang J, Boguski MS. Makałowski W, et al. Genome Res. 1996 Sep;6(9):846-57. doi: 10.1101/gr.6.9.846. Genome Res. 1996. PMID: 8889551 - Heterogeneous tempo and mode of conserved noncoding sequence evolution among four mammalian orders.
Babarinde IA, Saitou N. Babarinde IA, et al. Genome Biol Evol. 2013;5(12):2330-43. doi: 10.1093/gbe/evt177. Genome Biol Evol. 2013. PMID: 24259317 Free PMC article. - Molecular biology of type A endogenous retrovirus.
Ono M. Ono M. Kitasato Arch Exp Med. 1990 Sep;63(2-3):77-90. Kitasato Arch Exp Med. 1990. PMID: 1710682 Review. - Human and rodent DNA sequence comparisons: a mosaic model of genomic evolution.
Koop BF. Koop BF. Trends Genet. 1995 Sep;11(9):367-71. doi: 10.1016/s0168-9525(00)89108-8. Trends Genet. 1995. PMID: 7482789 Review.
Cited by
- Plant coilin: structural characteristics and RNA-binding properties.
Makarov V, Rakitina D, Protopopova A, Yaminsky I, Arutiunian A, Love AJ, Taliansky M, Kalinina N. Makarov V, et al. PLoS One. 2013;8(1):e53571. doi: 10.1371/journal.pone.0053571. Epub 2013 Jan 8. PLoS One. 2013. PMID: 23320094 Free PMC article. - Transcriptional regulation of translocator protein (Tspo) via a SINE B2-mediated natural antisense transcript in MA-10 Leydig cells.
Fan J, Papadopoulos V. Fan J, et al. Biol Reprod. 2012 May 10;86(5):147, 1-15. doi: 10.1095/biolreprod.111.097535. Print 2012 May. Biol Reprod. 2012. PMID: 22378763 Free PMC article. - Identification of a novel mutation in the PRCD gene causing autosomal recessive retinitis pigmentosa in a Turkish family.
Pach J, Kohl S, Gekeler F, Zobor D. Pach J, et al. Mol Vis. 2013 Jun 13;19:1350-5. Print 2013. Mol Vis. 2013. PMID: 23805042 Free PMC article. - Evolution of Codon Usage Bias in Diatoms.
Krasovec M, Filatov DA. Krasovec M, et al. Genes (Basel). 2019 Nov 6;10(11):894. doi: 10.3390/genes10110894. Genes (Basel). 2019. PMID: 31698749 Free PMC article. - Essential genes are more evolutionarily conserved than are nonessential genes in bacteria.
Jordan IK, Rogozin IB, Wolf YI, Koonin EV. Jordan IK, et al. Genome Res. 2002 Jun;12(6):962-8. doi: 10.1101/gr.87702. Genome Res. 2002. PMID: 12045149 Free PMC article.
References
- Bassett, D. E., Boguski, M. S. & Hieter, P. (1996) Nature (London) 589–590. - PubMed
- Li W-H. Molecular Evolution. Sunderland, MA: Sinauer; 1997.
- Hardison R C, Oeltjen J, Miller W. Genome Res. 1997;7:959–966. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases