Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli - PubMed (original) (raw)
Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli
B L Schneider et al. J Bacteriol. 1998 Aug.
Abstract
Arginine catabolism produces ammonia without transferring nitrogen to another compound, yet the only known pathway of arginine catabolism in Escherichia coli (through arginine decarboxylase) does not produce ammonia. Our aims were to find the ammonia-producing pathway of arginine catabolism in E. coli and to examine its function. We showed that the only previously described pathway of arginine catabolism, which does not produce ammonia, accounted for only 3% of the arginine consumed. A search for another arginine catabolic pathway led to discovery of the ammonia-producing arginine succinyltransferase (AST) pathway in E. coli. Nitrogen limitation induced this pathway in both E. coli and Klebsiella aerogenes, but the mechanisms of activation clearly differed in these two organisms. We identified the E. coli gene for succinylornithine aminotransferase, the third enzyme of the AST pathway, which appears to be the first of an astCADBE operon. Its disruption prevented arginine catabolism, impaired ornithine utilization, and affected the synthesis of all the enzymes of the AST pathway. Disruption of astB eliminated succinylarginine dihydrolase activity and prevented arginine utilization but did not impair ornithine catabolism. Overproduction of AST enzymes resulted in faster growth with arginine and aspartate. We conclude that the AST pathway is necessary for aerobic arginine catabolism in E. coli and that at least one enzyme of this pathway contributes to ornithine catabolism.
Figures
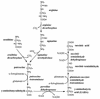
FIG. 1
Arginine catabolism by the ADC pathway and the routes of putrescine synthesis.
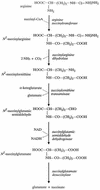
FIG. 2
AST pathway of arginine catabolism.
FIG. 3
Deduced amino acid sequence of succinylornithine transaminase. The underlined residues correspond to those determined by direct protein sequencing.
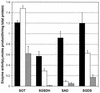
FIG. 4
AST enzyme activities in mutants with disruptions in astC and astB. Activity in W3110 (wild type) (black bars), in the astB derivative (white bars), and in the astC derivative (cross-hatched bars) is shown. The values given are the averages from at least five cultures. Activities are given in micromoles of product per hour per milligram of total protein; the error bars show the standard error of the mean. SOT, succinylornithine transaminase; SGSDH, succinylglutamic semialdehyde dehydrogenase; SAD, succinylarginine dihydrolase; SGDS, succinylglutamate desuccinylase.
Similar articles
- Cloning and characterization of the aru genes encoding enzymes of the catabolic arginine succinyltransferase pathway in Pseudomonas aeruginosa.
Itoh Y. Itoh Y. J Bacteriol. 1997 Dec;179(23):7280-90. doi: 10.1128/jb.179.23.7280-7290.1997. J Bacteriol. 1997. PMID: 9393691 Free PMC article. - N2-succinylornithine in ornithine catabolism of Pseudomonas aeruginosa.
Vander Wauven C, Jann A, Haas D, Leisinger T, Stalon V. Vander Wauven C, et al. Arch Microbiol. 1988;150(4):400-4. doi: 10.1007/BF00408314. Arch Microbiol. 1988. PMID: 3144259 - Regulation of polyamine biosynthesis by antizyme and some recent developments relating the induction of polyamine biosynthesis to cell growth. Review.
Canellakis ES, Kyriakidis DA, Rinehart CA Jr, Huang SC, Panagiotidis C, Fong WF. Canellakis ES, et al. Biosci Rep. 1985 Mar;5(3):189-204. doi: 10.1007/BF01119588. Biosci Rep. 1985. PMID: 3893559 Review. - Regulation of arginine biosynthesis, catabolism and transport in Escherichia coli.
Charlier D, Bervoets I. Charlier D, et al. Amino Acids. 2019 Aug;51(8):1103-1127. doi: 10.1007/s00726-019-02757-8. Epub 2019 Jul 3. Amino Acids. 2019. PMID: 31267155 Review.
Cited by
- Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125.
Médigue C, Krin E, Pascal G, Barbe V, Bernsel A, Bertin PN, Cheung F, Cruveiller S, D'Amico S, Duilio A, Fang G, Feller G, Ho C, Mangenot S, Marino G, Nilsson J, Parrilli E, Rocha EP, Rouy Z, Sekowska A, Tutino ML, Vallenet D, von Heijne G, Danchin A. Médigue C, et al. Genome Res. 2005 Oct;15(10):1325-35. doi: 10.1101/gr.4126905. Epub 2005 Sep 16. Genome Res. 2005. PMID: 16169927 Free PMC article. - P(II) signal transduction proteins, pivotal players in microbial nitrogen control.
Arcondéguy T, Jack R, Merrick M. Arcondéguy T, et al. Microbiol Mol Biol Rev. 2001 Mar;65(1):80-105. doi: 10.1128/MMBR.65.1.80-105.2001. Microbiol Mol Biol Rev. 2001. PMID: 11238986 Free PMC article. Review. - Transcriptional occlusion caused by overlapping promoters.
Zafar MA, Carabetta VJ, Mandel MJ, Silhavy TJ. Zafar MA, et al. Proc Natl Acad Sci U S A. 2014 Jan 28;111(4):1557-61. doi: 10.1073/pnas.1323413111. Epub 2014 Jan 13. Proc Natl Acad Sci U S A. 2014. PMID: 24474781 Free PMC article. - Effect of chemical chaperones in improving the solubility of recombinant proteins in Escherichia coli.
Prasad S, Khadatare PB, Roy I. Prasad S, et al. Appl Environ Microbiol. 2011 Jul;77(13):4603-9. doi: 10.1128/AEM.05259-11. Epub 2011 May 6. Appl Environ Microbiol. 2011. PMID: 21551288 Free PMC article. - Salmonella typhimurium nit is nadE: defective nitrogen utilization and ammonia-dependent NAD synthetase.
Schneider BL, Reitzer LJ. Schneider BL, et al. J Bacteriol. 1998 Sep;180(17):4739-41. doi: 10.1128/JB.180.17.4739-4741.1998. J Bacteriol. 1998. PMID: 9721319 Free PMC article.
References
- Bender, R. Personal communication.
- Clarke L, Carbon J. A colony bank containing synthetic Col E1 hybrid plasmids representative of the entire E. coli genome. Cell. 1976;9:91–99. - PubMed
- Dugan E L. Measurement of amino acids by column chromatography. Methods Enzymol. 1957;3:492–504.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases