trans-Complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells - PubMed (original) (raw)
trans-Complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells
A A Khromykh et al. J Virol. 1998 Sep.
Abstract
A BHK cell line persistently expressing a Kunjin (KUN) virus replicon RNA (repBHK, similar to our recently described ME/76Neo BHK cell line [A. A. Khromykh and E. G. Westaway, J. Virol. 71:1497-1505, 1997]) was used for rescue and propagation of KUN viruses defective in the RNA polymerase gene (NS5). A new infectious full-length KUN virus cDNA clone, FLSDX, prepared from our previously described cDNA clone pAKUN (A. A. Khromykh and E. G. Westaway, J. Virol. 68:4580-4588, 1994) and possessing approximately 10(5)-fold higher specific infectivity than that of pAKUN, was used for preparation of defective mutants. Deletions of the predicted RNA polymerase motif GDD (producing FLdGDD) and of one of the predicted methyltransferase motifs (S-adenosylmethionine [SAM] binding site, producing FLdSAM) were introduced separately into FLSDX. Transcription and transfection of FLdGDD and FLdSAM RNAs into repBHK cells but not into normal BHK cells resulted in their replication and the recovery of defective viruses able to replicate only in repBHK cells. Reverse transcription-PCR and sequencing analyses showed retention of the introduced deletions in the genomes of the recovered viruses. Retention of these deletions, as well as our inability to recover viruses able to replicate in normal BHK cells after prolonged incubation (for 7 days) of FLdGDD- or FLdSAM-transfected repBHK cells, excluded the possibility that recombination had occurred between the deleted defective NS5 genes present in transfected RNAs and the functional NS5 gene present in the repBHK cells. An RNA with a point mutation in the GDD motif (FLGVD) was also complemented in transfected repBHK cells, and defective virus was recovered by day 3 after transfection. However, in contrast to the results with FLdGDD and FLdSAM RNAs, prolonged (4 days or more) incubation of FLGVD RNA in normal BHK cells allowed recovery of a virus in which the GVD mutation had reverted via a single base change to the wild-type GDD sequence. Overall, these results represent the first demonstration of trans-complementation of defective flavivirus RNAs with deleterious deletions in the flavivirus RNA polymerase gene NS5. The complementation system described here may prove to be useful for the in vivo complementation of deletions and mutations affecting functional domains or the essential secondary structure in any of the other flavivirus nonstructural proteins.
Figures
FIG. 1
Construction and specific infectivities of the full-length KUN virus cDNA clones and the structures of KUN virus replicon RNAs. Schematic representations of the full-length constructs and of the constructs with deletions (replicon) show consecutive replacements of the cDNA fragments in the AKUN clone (stippled boxes) with analogous fragments obtained by RT-PCR from KUN virion RNA (shaded boxes) as described in Materials and Methods. PFU titers on the right-hand side of the figure are averages (of results from three experiments) obtained after electroporation of the transcribed RNAs into BHK21 cells and determined by plaque assay (see Materials and Methods); the titer of purified wild-type KUN virus RNA was ∼105 to ∼106 PFU/μg of RNA. Arrows marked Bgl(89), Sac(1481), Sph(2467), Dra(836), and Xho(11021) indicate restriction enzyme sites used in replacement cloning, with the numbers in parentheses representing nucleotide numbers in the KUN virus sequence (9, 14). An Expand High Fidelity PCR kit (Boehringer Mannheim) was used to obtain the indicated cDNA fragment of 6,895 nucleotides (nts) in the FLSD and FLSDX constructs, and Pfu PCR in FLSDX indicates that the cDNA fragment of 2,645 nucleotides was obtained with Pfu DNA polymerase (Stratagene). C20DXrep and C20DXrepNeo constructs were prepared as described in the Materials and Methods. Open boxes represent the deleted part of the genome (see reference 15). Ires, internal ribosomal entry site of encephalomyelitis virus RNA; Neo, neomycin transferase gene.
FIG. 2
Improvement of transfection efficacy of KUN virus replicon RNA and establishment of a replicon-expressing BHK cell line (repBHK) shown by IF analysis. (A and B) IF-positive BHK21 cells with anti-NS3 antibodies at 24 h after electroporation with the original C20rep RNA and with C20DXrep RNA of improved efficiency, respectively. (C) IF analysis with anti-NS3 antibodies of BHK cells transfected with C20DXNeo RNA (constructed as described in Materials and Methods) and maintained for 38 passages as repBHK cells in medium supplemented with 1 mg of G418 per ml, followed by nine passages in medium without G418. (D) IF analysis of repBHK cells (passage 33) with anti-E monoclonal antibodies. This figure and subsequent figures were prepared by scanning all the original data (slides, autoradiograms, etc.) on an Arcus II scanner (Agfa) with FotoLook software (Agfa) for a Macintosh computer at a resolution of 150 lines per in., followed by adjustment of the brightness and the contrast of some images, assembling of the montages with Microsoft PowerPoint 97 software, and printing of the images on an Epson Stylus Color 400 printer at a resolution of 720 dots per in. on Epson photo-quality ink-jet or Epson photo-quality glossy paper.
FIG. 3
Mutagenesis of KUN virus NS5 gene. (A) Schematic representation of the NS5 gene with motifs indicated; (B) Nucleotide and amino acid sequences of the region with a mutation and the regions with deletions. Numbers represent amino acid positions in the KUN virus NS5 gene (9). The filled box in panel A shows the boundaries of the region encompassing two proposed methyltransferase domains that include the SAM binding site (VIDLLGCGRGGW) (19), which is shown in boldface type in panel A. The hatched box in panel A shows boundaries of the region which includes a number of motifs proposed to be involved in RNA polymerase activity (3, 13, 18, 27, 28), including the RNA polymerase active site GDD shown in boldface type in panels A and B. R1 and F1 indicate the primers used for RT-PCR amplification (Fig. 4B). Dashed regions in panel B represent deleted nucleotides and corresponding amino acids. Boxed letters in panel B show new restriction sites introduced into the sequence during mutagenesis as described in Materials and Methods. (C) Autoradiogram of a sodium dodecyl sulfate–10% polyacrylamide gel containing electrophoresed samples of the [35S]methionine- and [35S]cysteine-labelled proteins translated in rabbit reticulocyte lysates programmed with the native and mutated NS5 RNA transcripts produced by T7 RNA polymerase from the intermediate plasmids containing the corresponding NS5 cDNA sequences in the pBluescript IIKS vector (see Materials and Methods). The arrow shows the position of the full-length NS5 protein. The KUN virus lane represents a [35S]methionine-labeled KUN virus-infected Vero cell lysate; dots identify NS5 and NS3. Numbers on the left represent Bio-Rad low-range prestained-protein standards. wt, wild type.
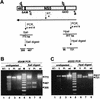
FIG. 4
Complementation in KUN virus replicon-expressing BHK (repBHK) cells of full-length KUN virus RNA with a GDD_-to-GVD mutation in the NS5 gene. (A) Northern blot analysis of total RNA isolated from repBHK cells (lanes 2 and 4) or normal BHK cells (lanes 3 and 5) at 3 days after transfection with FL_GVD RNA (p0) and at 2 days after infection with 3-day-posttransfection culture fluid (p1), with a radiolabelled cDNA probe representing 977 nucleotides of KUN virus prM and E genes (see Materials and Methods). Lane 1 contains mock-transfected repBHK cells, and lane 6 contains 10 ng of control FL_GVD_ RNA transcribed in vitro. An arrowhead indicates the position of RNA of about 11 kb, determined relative to migration in the same gels of ethidium bromide-stained λ DNA digested with Bst_EII (New England Biolabs). (B) RT-PCR analysis with F1 and R1 primers (Fig. 3A) of total RNA isolated from FL_GVD RNA-transfected normal BHK cells. Lane 1 contains λ DNA digested with Bst_EII (New England Biolabs). Lanes 2 and 3 contain RNA samples isolated from normal BHK cells at 3 and 6 days (3d and 6d), respectively, after electroporation with FL_GVD RNA. Lane 4 contains the control DNA fragment of 2.5 kb obtained by PCR amplification of FL_GVD_ cDNA. (C) Comparison of the nucleotide and deduced amino acid (boldface italic letters above the nucleotides) sequences in the GDD motif of wild-type (wt) and GVD mutated cDNAs with that of revertant (rev) cDNA. The sequence of the rev cDNA was obtained by automatic cycle sequencing of the purified 2.5-kb RT-PCR fragment shown in lane 3 in panel B with appropriate primers and by using an ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer, Brisbane, Australia). Boldface underlined letters represent nucleotides that either were targeted for mutation (
AT
in the wt sequence), mutated (
TC
in the GVD sequence), or reverted after replication of FL_GVD_ RNA in BHK cells (
AC
in the rev sequence). Boxed letters show the _Sal_I recognition site introduced by the GVD mutation.
FIG. 5
Complementation of full-length KUN virus RNA with a GDD deletion in the NS5 gene (construct FLd_GDD_). Results of IF analysis with anti-E antibodies (A) and Northern blot analysis with the E-specific probe (B) for detection of replicating FLd_GDD_ RNA are shown. Photos 1 and 2 in panel A and the corresponding repBHK lanes in panel B demonstrate replication of FLd_GDD_ RNA in repBHK cells at 3 and 5 days (3d and 5d), respectively, after electroporation. Photo 3 in panel A and the BHK lanes in panel B indicate the absence of replication of FLd_GDD_ RNA as late as 5 days after transfection into normal BHK cells. Photo 4 in panel A shows results of IF analysis with anti-E antibodies of repBHK cells at 2 days after infection with the complemented FLd_GDD_ virus recovered at 5 days after transfection of repBHK cells with FLd_GDD_ RNA. Photos 5 and 6 show results of dual-IF analysis with anti-E (fluorescein isothiocyanate [FITC] stain; photo 5) and anti-NS3 (Texas Red [TR] stain; photo 6) antibodies of normal BHK cells infected for 2 days with complemented FLd_GDD_ virus recovered at 5 days after transfection with FLd_GDD_ RNA. CF, culture fluid. The control lane in panel B contains 10 ng of in vitro-transcribed FLd_GDD_ RNA. The arrow indicates the position of RNA of about 11 kb, determined as described in the legend to Fig. 4. The Northern blot was exposed to X-ray film for 3 h.
FIG. 6
Complementation of full-length KUN virus RNA with a deletion of the SAM binding site in the NS5 gene (construct FLdSAM). Results of IF analysis with anti-E antibodies (A) and Northern blot analysis with the E-specific probe (B) for detection of replicating FLdSAM RNA are shown. Photos 1 to 3 in panel A and the corresponding repBHK lanes in panel B demonstrate replication of electroporated FLdSAM RNA in repBHK cells at 3, 5, and 7 days (3d, 5d, and 7d, respectively). BHK lanes in panel B show the absence of replication of FLdSAM RNA at 3, 5, and 7 days after transfection into normal BHK cells. Photo 4 in panel A shows results of IF analysis with anti-E antibodies of repBHK cells at 2 days after infection with complemented FLdSAM virus recovered at 7 days after transfection of repBHK cells with FLdSAM RNA. Photos 5 and 6 show results of dual-IF analysis with anti-E (fluorescein isothiocyanate [FITC] stain; photo 5) and anti-NS3 (Texas Red [TR] stain; photo 6) antibodies of normal BHK cells infected with complemented FLdSAM virus recovered at 7 days after transfection of repBHK cells with FLdSAM RNA and immunostained 2 days later. CF, culture fluid. The arrow in panel B indicates the position of RNA of about 11 kb, determined as described in the legend to Fig. 4. The Northern blot was exposed to X-ray film for 24 h, compared to 3 h for the blot in Fig. 5B.
FIG. 7
Determination of the structure of the defective genomes. (A) Schematic representation of the KUN virus genome in the vicinity of the NS5 gene and the details of the RT-PCR protocol. SAM and GDD represent deleted motifs (Fig. 3). a, b, c, and d represent primers used for RT and PCR and correspond to the published KUN virus plus-sense sequence (9, 14). a, nucleotides 10378 to 10398 (minus sense); b, nucleotides 9576 to 9597 (plus sense); c, nucleotides 8372 to 8400 (minus sense); d, nucleotides 7606 to 7621 (plus sense). Lines marked SalI and HpaI indicate the positions of new sites in the defective genomes introduced into their cDNAs during construction (Fig. 3B). Numbers indicate the predicted sizes of the fragments obtained by PCR amplification and restriction digestion. 4B, NS4B. (B and C) Results of RT-PCR and restriction digest analyses of the RNAs isolated from the culture fluid collected at 7 days after transfection of FLdSAM RNA and at 5 days after transfection of FLd_GDD_ RNA, respectively. The primer for RT was a for both reactions; the primer pairs for PCR were a and b for FLd_GDD_ samples and c and d for FLdSAM samples (see panel A). Lanes 1 in both panels B and C contain PCR products from the parallel control reaction lacking reverse transcriptase (RT−). Lanes 2 contain the PCR products obtained from the RT reactions performed with RNAs from the defective viruses (V) FLdSAM (B) and FLd_GDD_ (C). Lanes 3 contain the PCR products obtained after amplification of the plasmid DNAs (Pl) FLdSAM (B) and FLd_GDD_ (C). Lanes 4 contain the PCR products obtained from the parental FLSDX plasmid DNA with primers specific for SAM (B) and GDD (C). Lanes 6 and 7 contain restriction digests of the corresponding purified PCR fragments shown in lanes 2 to 4 with _Sal_I (B) or _Hpa_I (C) restrictases. Lanes 5 in both panels B and C contain a 100-bp molecular size marker (M) (Progene, Brisbane, Australia). Arrows show the sizes (in thousands) of the undigested and digested DNA fragments. wt, wild type.
Similar articles
- Efficient trans-complementation of the flavivirus kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase.
Khromykh AA, Sedlak PL, Guyatt KJ, Hall RA, Westaway EG. Khromykh AA, et al. J Virol. 1999 Dec;73(12):10272-80. doi: 10.1128/JVI.73.12.10272-10280.1999. J Virol. 1999. PMID: 10559344 Free PMC article. - trans-Complementation analysis of the flavivirus Kunjin ns5 gene reveals an essential role for translation of its N-terminal half in RNA replication.
Khromykh AA, Sedlak PL, Westaway EG. Khromykh AA, et al. J Virol. 1999 Nov;73(11):9247-55. doi: 10.1128/JVI.73.11.9247-9255.1999. J Virol. 1999. PMID: 10516033 Free PMC article. - Kunjin RNA replication and applications of Kunjin replicons.
Westaway EG, Mackenzie JM, Khromykh AA. Westaway EG, et al. Adv Virus Res. 2003;59:99-140. doi: 10.1016/s0065-3527(03)59004-2. Adv Virus Res. 2003. PMID: 14696328 Review. - A structural view of the RNA-dependent RNA polymerases from the Flavivirus genus.
Lu G, Gong P. Lu G, et al. Virus Res. 2017 Apr 15;234:34-43. doi: 10.1016/j.virusres.2017.01.020. Epub 2017 Jan 25. Virus Res. 2017. PMID: 28131854 Review.
Cited by
- Identification of Key Residues in Dengue Virus NS1 Protein That Are Essential for Its Secretion.
Tan BEK, Beard MR, Eyre NS. Tan BEK, et al. Viruses. 2023 Apr 30;15(5):1102. doi: 10.3390/v15051102. Viruses. 2023. PMID: 37243188 Free PMC article. - Zika Virus Replicates in the Vagina of Mice with Intact Interferon Signaling.
Lopez CA, Dulson SJ, Lazear HM. Lopez CA, et al. J Virol. 2022 Sep 28;96(18):e0121922. doi: 10.1128/jvi.01219-22. Epub 2022 Aug 30. J Virol. 2022. PMID: 36040178 Free PMC article. - Extended characterisation of five archival tick-borne viruses provides insights for virus discovery in Australian ticks.
O'Brien CA, Huang B, Warrilow D, Hazlewood JE, Bielefeldt-Ohmann H, Hall-Mendelin S, Pegg CL, Harrison JJ, Paramitha D, Newton ND, Schulz BL, Suhrbier A, Hobson-Peters J, Hall RA. O'Brien CA, et al. Parasit Vectors. 2022 Feb 18;15(1):59. doi: 10.1186/s13071-022-05176-z. Parasit Vectors. 2022. PMID: 35180893 Free PMC article. - A Putative Lipid-Associating Motif in the West Nile Virus NS4A Protein Is Required for Efficient Virus Replication.
Mikulasova A, Gillespie LK, Ambrose RL, Aktepe TE, Trenerry AM, Liebscher S, Mackenzie JM. Mikulasova A, et al. Front Cell Dev Biol. 2021 May 12;9:655606. doi: 10.3389/fcell.2021.655606. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34055786 Free PMC article. - Novel Flavivirus Attenuation Markers Identified in the Envelope Protein of Alfuy Virus.
Westlake D, Bielefeldt-Ohmann H, Prow NAA, Hall RAA. Westlake D, et al. Viruses. 2021 Jan 20;13(2):147. doi: 10.3390/v13020147. Viruses. 2021. PMID: 33498300 Free PMC article.
References
- Adams S C, Broom A K, Sammels L M, Hartnett A C, Howard M J, Coelen R J, Mackenzie J S, Hall R A. Glycosylation and antigenic variation among Kunjin virus isolates. Virology. 1995;206:49–56. - PubMed
- Bartholomeusz A I, Wright P J. Synthesis of dengue virus RNA in vitro: initiation and the involvement of proteins NS3 and NS5. Arch Virol. 1993;128:111–121. - PubMed
- Chu P W, Westaway E G. Replication strategy of Kunjin virus: evidence for recycling role of replicative form RNA as template in semiconservative and asymmetric replication. Virology. 1985;140:68–79. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials