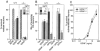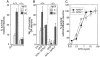GFRalpha1 is an essential receptor component for GDNF in the developing nervous system and kidney - PubMed (original) (raw)
doi: 10.1016/s0896-6273(00)80514-0.
I Fariñas, L C Wang, K Hagler, A Forgie, M Moore, M Armanini, H Phillips, A M Ryan, L F Reichardt, M Hynes, A Davies, A Rosenthal
Affiliations
- PMID: 9697851
- PMCID: PMC2710137
- DOI: 10.1016/s0896-6273(00)80514-0
GFRalpha1 is an essential receptor component for GDNF in the developing nervous system and kidney
G Cacalano et al. Neuron. 1998 Jul.
Abstract
Glial cell line-derived neurotrophic factor (GDNF) is a distant member of the TGFbeta protein family that is essential for neuronal survival and renal morphogenesis. We show that mice who are deficient in the glycosyl-phosphatidyl inositol (GPI) -linked protein GFRalpha1 (GDNFRalpha) display deficits in the kidneys, the enteric nervous system, and spinal motor and sensory neurons that are strikingly similar to those of the GDNF- and Ret-deficient mice. GFRalpha1-deficient dopaminergic and nodose sensory ganglia neurons no longer respond to GDNF or to the structurally related protein neurturin (NTN) but can be rescued when exposed to GDNF or neurturin in the presence of soluble GFRalpha1. In contrast, GFRalpha1-deficient submandibular parasympathetic neurons retain normal response to these two factors. Taken together with the available genetic and biochemical data, these findings support the idea that GFRalpha1 and the transmembrane tyrosine kinase Ret are both necessary receptor components for GDNF in the developing kidney and nervous system, and that GDNF and neurturin can mediate some of their activities through a second receptor.
Figures
Figure 1. Disruption of the GFRα1 Gene
(A) Targeting vector, wild-type GFRα1 allele and the disrupted allele. Amino acids 14–66 are missing from the disrupted gene. The location of the probe used in Southern blot is indicated (Probe). The directions of gene transcription are marked by horizontal arrows. (B) Detection of homologous recombination event in an ES clone by a Southern blot. (C) Genotype analysis of wild-type (+/+), heterozygous mutant (+/−), and homozygous mutant (−/−) animals by polymerase chain reaction. The band in the upper panel (GFRα1) is specific for the wild-type GFRα1 gene. The upper band in the lower panel (IL8R) represents a control fragment from the IL8 receptor gene. The lower band in this panel (neo) is specific for the neo gene. (D and E) In situ hybridization of wild-type (D) and _GFRα1_−/− (E) E15 mouse embryos with exon 2 GFRα1 probe. Abbreviations: drg, dorsal root ganglia; gut, gut; kid, kidney; sc, spinal cord; st, stomach; tg, trigeminal gangilon; vib, vibrissa; and vm, ventral midbrain. Scale bar, 1 mm.
Figure 2. Neuronal Populations in P0 wild-type (+/+) and _GFRα1_−/− (−/−) Mice
Tyrosine hydroxylase staining of substantia nigra (A and B), striatum (C and D), and locus coeruleus (E and F). Tyrosine hydroxylase is the rate-limiting enzyme in dopamine and noradrenaline synthesis. Cresyl violet staining of superior cervical ganglia (G and H) and petrosal nodose ganglia (I and J) neurons from 129 × CD-1, F2 mice. No deficits or abnormalities in neuronal number, morphology, or innervation pattern are detected in the _GFRα1_−/− mice. Scale bar: ~100 μm in (A), (B), (E), and (F), 30 μm in (C), (D), (I), and (J), and 50 μm in (G) and (H).
Figure 3. Enteric Nervous System in Wild-Type (+/+) and _GFRα1_−/− (−/−) Mice
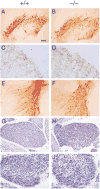
(A and B) Whole mounts of small intestine from E18 mice stained with the general neuronal antibody PGP 9.5. Inset in (A) depicts the expression of GFRα1 mRNA in E18 wild-type mouse gut as detected by in situ hybridization. (C–I) Section through the small intestine (C and D), colon (E and F), and rectum (G–I) of E17 embryos stained with neurofilament (C and D) or peripherin (E–I). Enteric neurons (ENS) were not found in the intestine and are very rarely found in the stomach and colon of the _GFRα1_−/− 129 × CD-1, F2 mice. No ENS neurons were detected in the colon of the _GDNF_−/− mice. (I) Sym represents afferent fibers probably derived from sympathetic innervation to the gut. Scale bar: ~100 μm in (A) and (B), 30 μm in (C) and (D), 50 μm in (E) and (F), and 300 μm in (G) through (J).
Figure 4. Kidneys in Wild-Type and _GFRα1_−/− Mice
(A and B) Photographs of the abdomen in E17 wild type (A) and _GDNF_−/− (B) 129 × CD-1, F2 mice. Note the position of the kidneys (Ki) subadjacent to the adrenals (Ad) in (A) and their absence in the mutant (B). (C–H) Sagittal sections through the kidney region of E12.5 wild-type (+/+) and _GFRα1_−/− (−/−) embryos stained with hematoxylin and eosin (C and D), Pax2 antibodies (E and F), or WT1 antibodies (G and H). Inset in (C) represents in situ hybridization of GFRα1 cDNA probe to developing nephrons and ureteric bud in wild-type kidney. Abbreviations: UB, uretric bud; MM, metanephric (condensing) mesenchyme; and NR, nephrogenic region (the region that undergoes mesenchymal-to-epithelial conversion and differentiated nephrons). Scale bar: ~500 μm in (A) and (B), 100 μm in (C) and (D), and 20 μm in (E) through (H).
Figure 5. Survival of Primary Embryonic Neurons from Wild-Type and _GFRα1_−/− Mice in the Presence of GDNF
The response of primary embryonic wild-type (+/+) and _GFRα1_−/− (−/−) nodose (A), dopaminergic (B), and submandibular (C) neurons from 129 × CD-1, F2 mice to GDNF. Neuronal survival is presented as percent or absolute number over control.
Figure 6. Survival of Primary Embryonic Neurons from Wild-Type and _GFRα1_−/− Mice in the Presence of NTN
The response of primary embryonic wild-type (+/+) and _GFRα1_−/− (−/−) nodose (A), dopaminergic (B), and submandibular (C) neurons from 129 × CD-1, F2 mice to NTN. Neuronal survival is presented as percent or absolute number over control.
Similar articles
- Expression of GDNF family receptor components during development: implications in the mechanisms of interaction.
Yu T, Scully S, Yu Y, Fox GM, Jing S, Zhou R. Yu T, et al. J Neurosci. 1998 Jun 15;18(12):4684-96. doi: 10.1523/JNEUROSCI.18-12-04684.1998. J Neurosci. 1998. PMID: 9614243 Free PMC article. - Broad specificity of GDNF family receptors GFRalpha1 and GFRalpha2 for GDNF and NTN in neurons and transfected cells.
Wang LC, Shih A, Hongo J, Devaux B, Hynes M. Wang LC, et al. J Neurosci Res. 2000 Jul 1;61(1):1-9. doi: 10.1002/1097-4547(20000701)61:1<1::AID-JNR1>3.0.CO;2-J. J Neurosci Res. 2000. PMID: 10861794 - Expression and regulation of GFRalpha3, a glial cell line-derived neurotrophic factor family receptor.
Naveilhan P, Baudet C, Mikaels A, Shen L, Westphal H, Ernfors P. Naveilhan P, et al. Proc Natl Acad Sci U S A. 1998 Feb 3;95(3):1295-300. doi: 10.1073/pnas.95.3.1295. Proc Natl Acad Sci U S A. 1998. PMID: 9448325 Free PMC article. - Other neurotrophic factors: glial cell line-derived neurotrophic factor (GDNF).
Saarma M, Sariola H. Saarma M, et al. Microsc Res Tech. 1999 May 15-Jun 1;45(4-5):292-302. doi: 10.1002/(SICI)1097-0029(19990515/01)45:4/5<292::AID-JEMT13>3.0.CO;2-8. Microsc Res Tech. 1999. PMID: 10383122 Review. - Novel functions and signalling pathways for GDNF.
Sariola H, Saarma M. Sariola H, et al. J Cell Sci. 2003 Oct 1;116(Pt 19):3855-62. doi: 10.1242/jcs.00786. J Cell Sci. 2003. PMID: 12953054 Review.
Cited by
- GDNF synthesis, signaling, and retrograde transport in motor neurons.
Cintrón-Colón AF, Almeida-Alves G, Boynton AM, Spitsbergen JM. Cintrón-Colón AF, et al. Cell Tissue Res. 2020 Oct;382(1):47-56. doi: 10.1007/s00441-020-03287-6. Epub 2020 Sep 8. Cell Tissue Res. 2020. PMID: 32897420 Free PMC article. Review. - Sympathoadrenal hyperplasia causes renal malformations in Ret(MEN2B)-transgenic mice.
Gestblom C, Sweetser DA, Doggett B, Kapur RP. Gestblom C, et al. Am J Pathol. 1999 Dec;155(6):2167-79. doi: 10.1016/S0002-9440(10)65534-4. Am J Pathol. 1999. PMID: 10595945 Free PMC article. - Brain-derived neurotrophic factor and glial cell line-derived neurotrophic factor are required simultaneously for survival of dopaminergic primary sensory neurons in vivo.
Erickson JT, Brosenitsch TA, Katz DM. Erickson JT, et al. J Neurosci. 2001 Jan 15;21(2):581-9. doi: 10.1523/JNEUROSCI.21-02-00581.2001. J Neurosci. 2001. PMID: 11160437 Free PMC article. - Ape1/Ref-1 Stimulates GDNF/GFRalpha1-mediated Downstream Signaling and Neuroblastoma Proliferation.
Kang MY, Kim KY, Yoon Y, Kang Y, Kim HB, Youn CK, Kim DH, Kim MH. Kang MY, et al. Korean J Physiol Pharmacol. 2009 Oct;13(5):349-56. doi: 10.4196/kjpp.2009.13.5.349. Epub 2009 Oct 31. Korean J Physiol Pharmacol. 2009. PMID: 19915696 Free PMC article. - Glial cell line-derived neurotrophic factor and target-dependent regulation of large-conductance KCa channels in developing chick lumbar motoneurons.
Martin-Caraballo M, Dryer SE. Martin-Caraballo M, et al. J Neurosci. 2002 Dec 1;22(23):10201-8. doi: 10.1523/JNEUROSCI.22-23-10201.2002. J Neurosci. 2002. PMID: 12451121 Free PMC article.
References
- Arenas E, Trupp M, Akerud P, Ibáñez CF. GDNF prevents degeneration and promotes the phenotype of brain noradrenergic neurons in vivo. Neuron. 1995;15:1465–1473. - PubMed
- Baloh RH, Tansey MG, Golden JP, Creedon DJ, Heuckeroth RO, Keck CL, Zimonjic DB, Popescu NC, Johnson EM, Milbrandt J. Trnr2, a novel receptor that mediates neurturin and GDNF signaling through Ret. Neuron. 1997;18:793–802. - PubMed
- Beck KD, Valverde J, Alexi T, Poulsen K, Moffat B, Vandlen RA, Rosenthal A, Hefti F. Mesencephalic dopaminergic neurons protected by GDNF from axotomy-induced degeneration in the adult brain. Nature. 1995;373:339–341. - PubMed
- Buj-Bello A, Buchman VL, Horton A, Rosenthal A, Davies AM. GDNF is an age-specific survival factor for sensory and autonomic neurons. Neuron. 1995;15:821–828. - PubMed
- Buj-Bello A, Adu J, Pinon L, Horton A, Thompson J, Rosenthal A, Chinchetru M, Buchman VL, Davies AM. Neurturin responsiveness requires a GPI-linked receptor and the Ret receptor tyrosine kinase. Nature. 1997;387:721–724. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials