A hypermutable insert in an immunoglobulin transgene contains hotspots of somatic mutation and sequences predicting highly stable structures in the RNA transcript - PubMed (original) (raw)
A hypermutable insert in an immunoglobulin transgene contains hotspots of somatic mutation and sequences predicting highly stable structures in the RNA transcript
U Storb et al. J Exp Med. 1998.
Abstract
Immunoglobulin (Ig) genes expressed in mature B lymphocytes can undergo somatic hypermutation upon cell interaction with antigen and T cells. The mutation mechanism had previously been shown to depend upon transcription initiation, suggesting that a mutator factor was loaded on an RNA polymerase initiating at the promoter and causing mutations during elongation (Peters, A., and U. Storb. 1996. Immunity. 4:57-65). To further elucidate this process we have created an artificial substrate consisting of alternating EcoRV and PvuII restriction enzyme sites (EPS) located within the variable (V) region of an Ig transgene. This substrate can easily be assayed for the presence of mutations in DNA from transgenic lymphocytes by amplifying the EPS insert and determining by restriction enzyme digestion whether any of the restriction sites have been altered. Surprisingly, the EPS insert was mutated many times more frequently than the flanking Ig sequences. In addition there were striking differences in mutability of the different nucleotides within the restriction sites. The data favor a model of somatic hypermutation where the fine specificity of the mutations is determined by nucleotide sequence preferences of a mutator factor, and where the general site of mutagenesis is determined by the pausing of the RNA polymerase due to secondary structures within the nascent RNA.
Figures
Figure 1
Maps of the EPS transgene and primers. (A) The EPS transgene and sequence of the EPS insert (not to scale). (B) The EPS transgene from the leader to the 5′ end of the J-C intron. Primers used for cloning and sequencing are shown below the transgene. (C) Maps of the EcoRV and PvuII sites within the EPS. Numbers indicate distances between the cut sites.
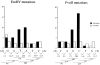
Figure 4
Relationship of mutations to hydrogen bonds and mutability of di- and trinucleotides within the EcoRV and PvuII sites. The columns show (black) the number of mutations from Fig. 3 in the EcoRV sites (EB to EG) and PvuII sites (PA to PF); (gray) the hydrogen bonds between the unmutated restriction site NTs G/C, C/G, A/T, and T/A base pairs. Below the graph are shown the mutability quotients (MQ; see Materials and Methods) of somatic mutations in di- and trinucleotides from a large database of normal Ig gene mutations (17). The numbers for dinucleotides are placed between two NTs in the EPS dinucleotide; the numbers for trinucleotides are placed below the central NT in the EPS trinucleotide. Bold numbers: the MQ is greatly higher than 1 (see legend Fig. 3). * Numbers: the MQ is greatly lower than 1. The first and last NT written in parentheses is the most frequent flanking NT. In the calculation of the MQ for given di- or trinucleotides of the first or last NTs, the MQs were averaged for all flanking NTs. The white boxes above the first G in the EcoRV site and the last G in the PvuII site indicate a NT that is overlapping both sites between PA and EB (see Fig. 1).
Figure 3
Mutations in the EPS and flanks. The original transgene sequence from NT 290–900 is shown in uppercase letters; below it in lower case letters are the observed mutations from all 46 sequences. The restriction sites within the EPS are indicated (E, EcoRV; P, PvuII; the seven Eco and six Pvu sites are labeled A–G and A–F, respectively). 3×, mutations in the EA site were found by restriction analysis in three clones that were lost before DNA sequencing could be performed; since in these clones the PA site was not mutated, the mutations can only be in position 1–5 of EA; the 6 mutations in EA are excluded from Fig. 4, but included in Fig. 6. The symbols below each sequence indicate the mutability quotient (MQ; see Materials and Methods) of the trinucleotide starting at the indicated NT (after reference 17). MQ symbols above the line: high column = very high MQ (≥1.44), low column = high MQ (1.36–1.43); MQ symbols below the line: high column = very low MQ (≤0.60), low column = low MQ (0.68–0.61); no symbol: MQ 0.69–1.35 (i.e., the observed mutability does not differ greatly from the expected mutability).
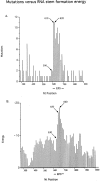
Figure 6
Mutations and RNA stem formation energies. (A) Mutations. NTs 290–900 from the start of the leader peptide sequence are shown (data are shown in Fig. 3). Numbers of point mutations in intervals of 10 nucleotides were summed. NTs 590 and 610/620 indicate the start and peak of the EPS related hypermutability, respectively. The EPS comprises nucleotides 608–703. (B) Energy. An example of a predicted RNA secondary structure and its folding energy is shown in Fig. 5. The RNA folding energies (expressed as negative numbers) were determined for the transgene sequence from position 290–900 for a window of 50 nucleotides at 5 nucleotide intervals. For the plot, energies of 10 nt intervals were derived by averaging the energies from three readings of the most 3′ positions within ±5 nucleotides of the plotted position. The start and peak of the EPS-related energy is indicated as 630 and 650/660, respectively.
Figure 5
One example of the determination of the predicted energy of RNA folding in a 50 NT window from a reiterative scanning of the Mfold program. The nascent RNA is reportedly paired with the transcribed DNA for 2–12 NTs (26), or 9–10 NTs (27). Upstream of this DNA/RNA hybrid a potential RNA double stem of high predicted stability may arrest the elongating RNA polymerase located at the 3′ end of the nascent RNA (see Fig. 7).
Figure 7
Model of somatic hypermutation of Ig genes. MuF, mutator factor; E, Ig enhancer; IEBP, Ig enhancer binding proteins; pol, RNA polymerase II; exo, DNA exonuclease; x, point mutation; Fen1, flap endonuclease 1.
Figure 2
PAGE of EPS PCR products from 10 clones digested with PvuII or EcoRV. The EPS in B1 and A1 is not mutated. P, PvuII; E, EcoRV.
Similar articles
- Somatic hypermutation of an artificial test substrate within an Ig kappa transgene.
Klotz EL, Hackett J Jr, Storb U. Klotz EL, et al. J Immunol. 1998 Jul 15;161(2):782-90. J Immunol. 1998. PMID: 9670955 - Cis-acting sequences that affect somatic hypermutation of Ig genes.
Storb U, Peters A, Klotz E, Kim N, Shen HM, Hackett J, Rogerson B, Martin TE. Storb U, et al. Immunol Rev. 1998 Apr;162:153-60. doi: 10.1111/j.1600-065x.1998.tb01438.x. Immunol Rev. 1998. PMID: 9602361 Review. - Strand bias in Ig somatic hypermutation is determined by signal sequence within the variable region.
Ching AK, Li PS, Chan WY, Ma CH, Lee SS, Lim PL, Chui YL. Ching AK, et al. Int Immunol. 2000 Sep;12(9):1245-53. doi: 10.1093/intimm/12.9.1245. Int Immunol. 2000. PMID: 10967019 - Immunoglobulin transgenes as targets for somatic hypermutation.
Storb U, Peters A, Klotz E, Kim N, Shen HM, Hackett J, Rogerson B, O'Brien R, Martin TE. Storb U, et al. Int J Dev Biol. 1998;42(7):977-82. Int J Dev Biol. 1998. PMID: 9853828 Review. - Different mismatch repair deficiencies all have the same effects on somatic hypermutation: intact primary mechanism accompanied by secondary modifications.
Kim N, Bozek G, Lo JC, Storb U. Kim N, et al. J Exp Med. 1999 Jul 5;190(1):21-30. doi: 10.1084/jem.190.1.21. J Exp Med. 1999. PMID: 10429667 Free PMC article.
Cited by
- Generation and repair of AID-initiated DNA lesions in B lymphocytes.
Chen Z, Wang JH. Chen Z, et al. Front Med. 2014 Jun;8(2):201-16. doi: 10.1007/s11684-014-0324-4. Epub 2014 Apr 21. Front Med. 2014. PMID: 24748462 Free PMC article. Review. - The role of activation-induced deaminase in antibody diversification and genomic instability.
Wang JH. Wang JH. Immunol Res. 2013 Mar;55(1-3):287-97. doi: 10.1007/s12026-012-8369-4. Immunol Res. 2013. PMID: 22956489 Review. - Hypermutation in derepressed operons of Escherichia coli K12.
Wright BE, Longacre A, Reimers JM. Wright BE, et al. Proc Natl Acad Sci U S A. 1999 Apr 27;96(9):5089-94. doi: 10.1073/pnas.96.9.5089. Proc Natl Acad Sci U S A. 1999. PMID: 10220423 Free PMC article. - Transcriptional pausing and stalling causes multiple clustered mutations by human activation-induced deaminase.
Canugovi C, Samaranayake M, Bhagwat AS. Canugovi C, et al. FASEB J. 2009 Jan;23(1):34-44. doi: 10.1096/fj.08-115352. Epub 2008 Sep 4. FASEB J. 2009. PMID: 18772346 Free PMC article. - Variable deletion and duplication at recombination junction ends: implication for staggered double-strand cleavage in class-switch recombination.
Chen X, Kinoshita K, Honjo T. Chen X, et al. Proc Natl Acad Sci U S A. 2001 Nov 20;98(24):13860-5. doi: 10.1073/pnas.241524898. Proc Natl Acad Sci U S A. 2001. PMID: 11717442 Free PMC article.
References
- Kelsoe G. The germinal center reaction. Immunol Today. 1995;16:324–326. - PubMed
- MacLennan I. Germinal centers. Annu Rev Immunol. 1994;112:117–139. - PubMed
- Storb U. The molecular basis of somatic hypermutation of immunoglobulin genes. Curr Opin Immunol. 1996;8:206–214. - PubMed
- Betz A, Milstein C, Gonzalez-Fernandes R, Pannell R, Larson T, Neuberger M. Elements regulating somatic hypermutation of an immunoglobulin κ gene: critical role for the intron enhancer/matrix attachment region. Cell. 1994;77:239–248. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI07090/AI/NIAID NIH HHS/United States
- GM07183/GM/NIGMS NIH HHS/United States
- T32 AI007090/AI/NIAID NIH HHS/United States
- GM38649/GM/NIGMS NIH HHS/United States
- T32 GM007183/GM/NIGMS NIH HHS/United States