The early stage of folding of villin headpiece subdomain observed in a 200-nanosecond fully solvated molecular dynamics simulation - PubMed (original) (raw)
The early stage of folding of villin headpiece subdomain observed in a 200-nanosecond fully solvated molecular dynamics simulation
Y Duan et al. Proc Natl Acad Sci U S A. 1998.
Abstract
A new approach in implementing classical molecular dynamics simulation for parallel computers has enabled a simulation to be carried out on a protein with explicit representation of water an order of magnitude longer than previously reported and will soon enable such simulations to be carried into the microsecond time range. We have used this approach to study the folding of the villin headpiece subdomain, a 36-residue small protein consisting of three helices, from an unfolded structure to a molten globule state, which has a number of features of the native structure. The time development of the solvation free energy, the radius of gyration, and the mainchain rms difference from the native NMR structure showed that the process can be seen as a 60-nsec "burst" phase followed by a slow "conformational readjustment" phase. We found that the burial of the hydrophobic surface dominated the early phase of the folding process and appeared to be the primary driving force of the reduction in the radius of gyration in that phase.
Figures
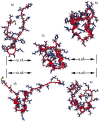
Figure 1
Ribbon representations of the unfolded (a) and the native (c) structures, and the snapshots at 85 nsec (b), at 104 nsec (d), and at 182 nsec (e), generated by using University of California at San Francisco’s
midasplus
. rmsds from the native state are given in the figure.
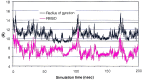
Figure 2
Rγ and the mainchain rmsd from the native structure as a function of time. The straight lines represent the Rγ of the starting structure (upper line) and the native structure (lower line).
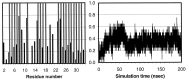
Figure 3
Helical content measured by the mainchain φ-ψ angle (−60 ± 30, −40 ± 30) from 10,000 snapshots (20 psec intervals). (Left) The average over the trajectory. The shaded bars represent the residues that are helical in the NMR structure. (Right) The fractional native helical content (i.e., those presented in both the native and the simulated structures divided by the total in the native structure) as a function of time.
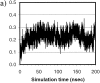
Figure 4
(a) Fractional native contacts, Q, as a function of time. The native contacts were measured as the number of neighboring residues presented in the native structure. Residues are taken to be in contact if any of the atom pairs (including both sidechain and mainchain atoms) are closer than 2.8 Å, excluding residues i and i+1, which always have the contacts through mainchain atoms. Fractional native contact is the number of total native contacts presented in the simulated structure divided by the number of native contacts in the native structure. (b) SFE of the protein as a function of time. The upper dashed line represents the SFE of the starting structure, and the lower dotted line represents that of the native structure. The parameters are those by Eisenberg and McLachlan (43), (i.e., 0.0163, −0.00637, 0.02114, −0.02376, −0.05041, in kcal/mole/Å2, for the surface areas of nonpolar, polar, sulfur, charged oxygen, and charged nitrogen, respectively).
Figure 4
(a) Fractional native contacts, Q, as a function of time. The native contacts were measured as the number of neighboring residues presented in the native structure. Residues are taken to be in contact if any of the atom pairs (including both sidechain and mainchain atoms) are closer than 2.8 Å, excluding residues i and i+1, which always have the contacts through mainchain atoms. Fractional native contact is the number of total native contacts presented in the simulated structure divided by the number of native contacts in the native structure. (b) SFE of the protein as a function of time. The upper dashed line represents the SFE of the starting structure, and the lower dotted line represents that of the native structure. The parameters are those by Eisenberg and McLachlan (43), (i.e., 0.0163, −0.00637, 0.02114, −0.02376, −0.05041, in kcal/mole/Å2, for the surface areas of nonpolar, polar, sulfur, charged oxygen, and charged nitrogen, respectively).
Similar articles
- Pathways to a protein folding intermediate observed in a 1-microsecond simulation in aqueous solution.
Duan Y, Kollman PA. Duan Y, et al. Science. 1998 Oct 23;282(5389):740-4. doi: 10.1126/science.282.5389.740. Science. 1998. PMID: 9784131 - All-atom fast protein folding simulations: the villin headpiece.
Shen MY, Freed KF. Shen MY, et al. Proteins. 2002 Dec 1;49(4):439-45. doi: 10.1002/prot.10230. Proteins. 2002. PMID: 12402354 - Experimental tests of villin subdomain folding simulations.
Kubelka J, Eaton WA, Hofrichter J. Kubelka J, et al. J Mol Biol. 2003 Jun 13;329(4):625-30. doi: 10.1016/s0022-2836(03)00519-9. J Mol Biol. 2003. PMID: 12787664 - Combining experiment and simulation in protein folding: closing the gap for small model systems.
Schaeffer RD, Fersht A, Daggett V. Schaeffer RD, et al. Curr Opin Struct Biol. 2008 Feb;18(1):4-9. doi: 10.1016/j.sbi.2007.11.007. Epub 2008 Feb 1. Curr Opin Struct Biol. 2008. PMID: 18242977 Free PMC article. Review. - Membrane protein folding.
Booth PJ, Curran AR. Booth PJ, et al. Curr Opin Struct Biol. 1999 Feb;9(1):115-21. doi: 10.1016/s0959-440x(99)80015-3. Curr Opin Struct Biol. 1999. PMID: 10047580 Review.
Cited by
- Protein storytelling through physics.
Brini E, Simmerling C, Dill K. Brini E, et al. Science. 2020 Nov 27;370(6520):eaaz3041. doi: 10.1126/science.aaz3041. Science. 2020. PMID: 33243857 Free PMC article. Review. - Enhanced Thermostability and Enzymatic Activity of Cel6A Variants from Thermobifida fusca by Empirical Domain Engineering (Short Title: Domain Engineering of Cel6A).
Ali I, Rehman HM, Mirza MU, Akhtar MW, Asghar R, Tariq M, Ahmed R, Tanveer F, Khalid H, Alghamdi HA, Froeyen M. Ali I, et al. Biology (Basel). 2020 Aug 7;9(8):214. doi: 10.3390/biology9080214. Biology (Basel). 2020. PMID: 32784797 Free PMC article. - Limitations and challenges in protein stability prediction upon genome variations: towards future applications in precision medicine.
Sanavia T, Birolo G, Montanucci L, Turina P, Capriotti E, Fariselli P. Sanavia T, et al. Comput Struct Biotechnol J. 2020 Jul 24;18:1968-1979. doi: 10.1016/j.csbj.2020.07.011. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 32774791 Free PMC article. Review. - Studying the role of cooperative hydration in stabilizing folded protein states.
Huggins DJ. Huggins DJ. J Struct Biol. 2016 Dec;196(3):394-406. doi: 10.1016/j.jsb.2016.09.003. Epub 2016 Sep 12. J Struct Biol. 2016. PMID: 27633532 Free PMC article. - Characterizing a partially ordered miniprotein through folding molecular dynamics simulations: Comparison with the experimental data.
Baltzis AS, Glykos NM. Baltzis AS, et al. Protein Sci. 2016 Mar;25(3):587-96. doi: 10.1002/pro.2850. Epub 2015 Dec 16. Protein Sci. 2016. PMID: 26609791 Free PMC article.
References
- Sifers R N. Nat Struct Biol. 1995;2:355–357. - PubMed
- Prusiner S B. Science. 1997;278:245–251. - PubMed
- Skolnick J, Kolinski A. Science. 1990;250:1121–1125. - PubMed
- Sali A, Shakhnovich E, Karplus M. Nature (London) 1994;369:248–251. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources