Distribution of genes in the genome of Arabidopsis thaliana and its implications for the genome organization of plants - PubMed (original) (raw)
Comparative Study
Distribution of genes in the genome of Arabidopsis thaliana and its implications for the genome organization of plants
A Barakat et al. Proc Natl Acad Sci U S A. 1998.
Abstract
Previous work has shown that, in the large genomes of three Gramineae [rice, maize, and barley: 415, 2,500, and 5,300 megabases (Mb), respectively] most genes are clustered in long DNA segments (collectively called the "gene space") that represent a small fraction (12-24%) of nuclear DNA, cover a very narrow (0.8-1.6%) GC range, and are separated by vast expanses of gene-empty sequences. In the present work, we have analyzed the small (ca. 120 Mb) nuclear genome of Arabidopsis thaliana and shown that its organization is drastically different from that of the genomes of Gramineae. Indeed, (i) genes are distributed over about 85% of the main band of DNA in CsCl and cover an 8% GC range; (ii) ORFs are fairly evenly distributed in long (>50 kb) sequences from GenBank that amount to about 10 Mb; and (iii) the GC levels of protein-coding sequences (and of their third codon positions) are correlated with the GC levels of their flanking sequences. The different pattern of gene distribution of Arabidopsis compared with Gramineae appears to be because the genomes of the latter comprise (i) many large gene-empty regions separating gene clusters and (ii) abundant transposons in the intergenic sequences of gene clusters. Both sequences are absent or very scarce in the Arabidopsis genome. These observations provide a comparative view of angiosperm genome organization.
Figures
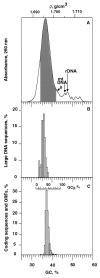
Figure 1
(A) Absorbance profile of Arabidopsis nuclear DNA as obtained by centrifugation in a CsCl analytical density gradient. The shoulder (s) may correspond to contaminating chloroplast DNA, the following small peaks to contaminating mitochondrial DNA (ρ = 1.706 g/cm3), rDNA (ρ = 1.707 g/cm3), and to three satellite DNAs (see text). The shaded area corresponds to the DNA fractions containing nuclear protein-encoding genes (see legend of Fig. 2). (B) Compositional distribution of large (>50 kb) GenBank DNA sequences from Arabidopsis. (C) Gene distribution obtained by plotting the relative number of Arabidopsis genes against their GC3 values (top scale); 2,490 sequences from GenBank (release 103; October 15, 1997) were used to construct the histogram. In C, the common GC abscissa of the three plots represents the GC values of the DNA fractions containing the genes (as derived from Fig. 2).
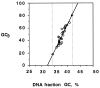
Figure 2
Plot of GC3 of genes (circles) versus GC values of DNA fractions corresponding to the hybridization peaks (from the data of Table 1). The solid circles represent the two extreme GC3 values of Arabidopsis genes as found in GenBank. The vertical broken lines indicate the GC range of the DNA fractions containing the genes. This was used to define in Fig. 1_A_ (shaded area) the DNA range in which genes are located.
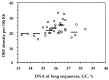
Figure 3
ORF density (number of ORFs per 100 kb) in large (>50 kb) DNA segments from Arabidopsis (circles). Average values were also estimated for each 1% GC bin (horizontal bars).
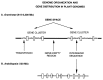
Figure 4
A scheme of genome organization and gene distribution in plant genomes. (A) In the large genomes of Gramineae, genes (large vertical boxes) are present in long gene clusters, which are separated from each other by gene-empty regions formed by repeated sequences (thick solid line). The ensemble of gene clusters forms the gene space. The intergenic sequences are compositionally very homogenous because largely formed by transposons (small horizontal boxes in the intergenic sequences). (B) The small genome of Arabidopsis essentially differs from the genomes of Gramineae because of (i) the disappearance (or very strong reduction) of gene-empty regions; (ii) the practical absence of transposons in intergenic sequences; and (iii) the higher gene density.
Similar articles
- The distribution of T-DNA in the genomes of transgenic Arabidopsis and rice.
Barakat A, Gallois P, Raynal M, Mestre-Ortega D, Sallaud C, Guiderdoni E, Delseny M, Bernardi G. Barakat A, et al. FEBS Lett. 2000 Apr 14;471(2-3):161-4. doi: 10.1016/s0014-5793(00)01393-4. FEBS Lett. 2000. PMID: 10767414 - The distribution of genes in the genomes of Gramineae.
Barakat A, Carels N, Bernardi G. Barakat A, et al. Proc Natl Acad Sci U S A. 1997 Jun 24;94(13):6857-61. doi: 10.1073/pnas.94.13.6857. Proc Natl Acad Sci U S A. 1997. PMID: 9192656 Free PMC article. - The gene distribution in the genomes of pea, tomato and date palm.
Barakat A, Han DT, Benslimane A, Rode A, Bernardi G. Barakat A, et al. FEBS Lett. 1999 Dec 10;463(1-2):139-42. doi: 10.1016/s0014-5793(99)01587-2. FEBS Lett. 1999. PMID: 10601654 - Colinearity and gene density in grass genomes.
Keller B, Feuillet C. Keller B, et al. Trends Plant Sci. 2000 Jun;5(6):246-51. doi: 10.1016/s1360-1385(00)01629-0. Trends Plant Sci. 2000. PMID: 10838615 Review. - Angiosperm mitochondrial genomes and mutations.
Kubo T, Newton KJ. Kubo T, et al. Mitochondrion. 2008 Jan;8(1):5-14. doi: 10.1016/j.mito.2007.10.006. Epub 2007 Nov 4. Mitochondrion. 2008. PMID: 18065297 Review.
Cited by
- The tomato genome: implications for plant breeding, genomics and evolution.
Ranjan A, Ichihashi Y, Sinha NR. Ranjan A, et al. Genome Biol. 2012 Aug 30;13(8):167. doi: 10.1186/gb-2012-13-8-167. Genome Biol. 2012. PMID: 22943138 Free PMC article. - Genomic organization of the S locus: Identification and characterization of genes in SLG/SRK region of S(9) haplotype of Brassica campestris (syn. rapa).
Suzuki G, Kai N, Hirose T, Fukui K, Nishio T, Takayama S, Isogai A, Watanabe M, Hinata K. Suzuki G, et al. Genetics. 1999 Sep;153(1):391-400. doi: 10.1093/genetics/153.1.391. Genetics. 1999. PMID: 10471721 Free PMC article. - The GC-heterogeneity of teleost fishes.
Melodelima C, Gautier C. Melodelima C, et al. BMC Genomics. 2008 Dec 24;9:632. doi: 10.1186/1471-2164-9-632. BMC Genomics. 2008. PMID: 19108743 Free PMC article. - Large-scale SNP discovery through RNA sequencing and SNP genotyping by targeted enrichment sequencing in cassava (Manihot esculenta Crantz).
Pootakham W, Shearman JR, Ruang-Areerate P, Sonthirod C, Sangsrakru D, Jomchai N, Yoocha T, Triwitayakorn K, Tragoonrung S, Tangphatsornruang S. Pootakham W, et al. PLoS One. 2014 Dec 31;9(12):e116028. doi: 10.1371/journal.pone.0116028. eCollection 2014. PLoS One. 2014. PMID: 25551642 Free PMC article. - Patterns and evolution of nucleotide landscapes in seed plants.
Serres-Giardi L, Belkhir K, David J, Glémin S. Serres-Giardi L, et al. Plant Cell. 2012 Apr;24(4):1379-97. doi: 10.1105/tpc.111.093674. Epub 2012 Apr 6. Plant Cell. 2012. PMID: 22492812 Free PMC article.
References
- Salinas J, Matassi G, Montero L M, Bernardi G. Nucleic Acids Res. 1988;19:5561–5567. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous