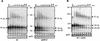Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant - PubMed (original) (raw)
Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant
R Boeck et al. Mol Cell Biol. 1998 Sep.
Abstract
mRNA in the yeast Saccharomyces cerevisiae is primarily degraded through a pathway that is stimulated by removal of the mRNA cap structure. Here we report that a mutation in the SPB8 (YJL124c) gene, initially identified as a suppressor mutation of a poly(A)-binding protein (PAB1) gene deletion, stabilizes the mRNA cap structure. Specifically, we find that the spb8-2 mutation results in the accumulation of capped, poly(A)-deficient mRNAs. The presence of this mutation also allows for the detection of mRNA species trimmed from the 3' end. These data show that this Sm-like protein family member is involved in the process of mRNA decapping, and they provide an example of 3'-5' mRNA degradation intermediates in yeast.
Figures
FIG. 1
Identification of YJL124c as the SPB8 gene. (A) Predicted open reading frame of Spb8p. The Sm-like region of the protein that is homologous to the Sm proteins is boxed. (B) Diagram of the YJL124c gene and the sites of mutations in the spb8-1 and spb8-2 alleles. spb8-1 contains a frameshift mutation at codon 145; spb8-2 contains a mini-Tn_3_ insertion within codon 32. (C) Sequence alignment of Spb8p with other Sm proteins based on an Sm domain alignment previously described (38). Database accession numbers for protein sequences: SmB (human), S10594; SmE (human), P08578; and SmX2 (alfalfa), P24715. SmX7, Brassica campestris pekinensis sequence derived by translation from nucleic acid sequence L33514.
FIG. 2
Complementation of the spb8 phenotype by wild-type SPB8. Yeast cells (YAS2266 to YAS2271) containing a deletion of PAB1 and the indicated allele of SPB8 in the genome, PAB1 on a URA3 CEN plasmid, and a TRP1 CEN plasmid with either SPB8 or no insert were streaked onto minimal medium lacking tryptophan and containing 1 mg of 5-FOA per ml (16) and grown at 30°C for 8 days. A photograph of the plate is shown.
FIG. 3
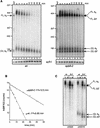
Degradation of MFA2pG mRNA in the spb8-2 strain. (A) Accumulation of MFA2pG mRNA degradation intermediates. Yeast strains YAS2272 (SPB8) and YAS2273 (spb8-2) carrying the GAL1:MFA2pG reporter (pRP485) (9) were grown in galactose-containing minimal medium. Glucose was then added to repress transcription of the reporter. RNA was recovered from cells harvested at the indicated times after repression, separated on a 6% polyacrylamide gel, and detected by hybridization with oligo(C) probe oRP121 (26). The length and mobility of size standards are indicated in nucleotides to the left of each gel. The lower panel shows the hybridization signal with the SCR1 probe, which recognizes a polymerase III transcript (12) that serves as an internal loading standard for these experiments (9). The lane containing the RNA sample treated with RNase H-oligo(dT) (0dT) serves to provide size markers for the deadenylated mRNA species. Positions of the full-length and oligo(G) deadenylated mRNAs [FL A0 and (G) A0] and the shorter fragments [FL Δ3′ and (G) Δ3′] are indicated. wt, wild type. (B) MFA2 mRNA is stabilized in an spb8-2 strain. The level of full-length MFA2 mRNA in each lane shown in panel A was quantified by phosphorimaging and then plotted as a function of time. The plotted values have been normalized to an SCR1 loading control. t1/2, half-life. (C) The shortened oligo(G) degradation intermediates in the spb8-2 strain lack mRNA sequences 5′ to the oligo(G) tract. mRNA samples derived from the 0- and 30-min time points in panel A were resolved on a 6% polyacrylamide gel and detected by Northern blot analysis with end-labeled oligonucleotides complementary to a region ending 19 nt 5′ to the oligo(G) tract (oAS22) or starting 13 nt 3′ to the oligo(G) tract (oAS318). Positions of the full-length and oligo(G) deadenylated mRNAs [FL A0 and (G) A0] and the shorter fragments [FL Δ3′ and (G) Δ3′] are indicated.
FIG. 4
Degradation of PGK1pG mRNA in the spb8-2 strain. (A) Accumulation of shortened PGK1pG mRNA species in an spb8-2 strain. Yeast strains YAS2274 (SPB8) and YAS2275 (spb8-2) carrying the GAL1:PGK1pG reporter (pRP602) (27) were grown in galactose-containing medium. RNA transcriptional shutoff and subsequent analysis were as for Fig. 3A except that all RNA samples were treated with RNase H-oRP70 in order to resolve the full-length PGK1pG molecules. Positions of the full-length and oligo(G) deadenylated mRNAs [FL A0 and (G) A0] and the oligo(G) shorter fragment [(G) Δ3′] are indicated. wt, wild type. (B) Cycloheximide induces the appearance of shortened PGK1pG mRNA species in a wild-type strain. Yeast strain YAS2274 (SPB8) carrying the GAL1:PGK1pG reporter was grown in galactose-containing medium and shifted to glucose medium containing 100 μg of cycloheximide (cyclo) per ml. RNA samples were analyzed as for Fig. 3A. Positions of the full-length and oligo(G) deadenylated mRNAs [FL A0 and (G) A0] and the oligo(G) shorter fragment [(G) Δ3′] are indicated. Sizes are indicated in nucleotides.
FIG. 5
Accumulation of deadenylated CUP1 mRNA in spb8-2 strains. Yeast strains YAS1915 (SPB8) (wild type [wt]) and YAS2278 (spb8-2) were grown to mid-log phase in YM medium and then induced for CUP1 synthesis by the addition of 0.5 mM CuSO4 to the medium. Aliquots of cells were taken at the indicated time points after induction; the mRNA within them was extracted, resolved on 6% polyacrylamide gel, and analyzed by Northern blot analysis with a CUP1 probe. Positions of the four deadenylated CUP1 transcripts containing alternative 5′ ends are indicated with arrows. Sizes are indicated in nucleotides. 40dT, 40-min time point for RNase H-oligo(dT) treatment.
FIG. 6
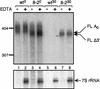
Stabilization of capped, deadenylated mRNA in the wild-type (wt) and spb8-2 (8-2) mutant strains. RNA samples derived from the zero (wt0 or 8-20) or 30-min (wt30 or 8-230) time points of the experiments shown in Fig. 2 were subject to treatment with 400 ng of purified Xrn1p in the presence (+) or absence (−) of neutralizing EDTA. Following resolution of the RNA samples on a 6% polyacrylamide gel, the MFA2pG mRNA was detected with an oligo(C) probe (oRP121). The 7S pre-rRNA was detected with an end-labeled oligonucleotide probe (oAS321) that specifically recognizes the 7S rRNA precursor. Positions of the full-length deadenylated mRNA (FL A0) and the shorter fragments (FL Δ3′) are indicated. Sizes are indicated in nucleotides.
FIG. 7
The shortened mRNA species in the spb8-2 mutant arise in wild-type yeast cells treated with cycloheximide and are not polyadenylated in _pab1_Δ strains. (A) Accumulation of shortened MFA2pG mRNA in a wild-type (wt) strain treated with cycloheximide (cyclo). Yeast strains YAS2272 (SPB8) and YAS2273 (spb8-2) carrying the GAL1:MFA2pG reporter were grown in galactose-containing medium and shifted to glucose medium to shut off transcription of the MFA2 mRNA. When indicated, cycloheximide was added at time zero of transcriptional shutoff. RNA samples derived from cells harvested at the indicated times after glucose addition were resolved on 6% polyacrylamide gels, and the MFA2 mRNA was detected by hybridization to end-labeled oligo(C). The length and mobility of size standards are indicated in nucleotides at the left. The lower panel shows the hybridization signal with the SCR1 probe. The lanes containing the RNA samples treated with RNase H-oligo(dT) (0dT and 40dT) provide size markers for the deadenylated mRNA species. The position of the full-length and oligo(G) deadenylated mRNAs [FL A0 and (G) A0] and the shorter fragments [FL Δ3′ and (G) Δ3′] are indicated. (B) The shortened MFA2pG mRNA intermediate is not derived from a polyadenylated precursor. Yeast strain YAS2279 (_spb8-2 pab1_Δ) carrying the GAL1:MFA2pG reporter and growing in galactose was harvested, and its MFA2pG mRNA was detected by Northern blot analysis with either prior treatment (0dT) or no treatment (0) with RNase H-oligo(dT) to remove the poly(A) tail. Positions of the full-length and oligo(G) deadenylated mRNAs [FL A0 and (G) A0] and the shorter oligo(G) fragment [(G) Δ3′] are indicated. The MFA2 mRNA was detected by hybridization to end-labeled oligo(C).
FIG. 8
The spb8-2 mutation does not alter the stability of a nonsense codon containing mRNA. mRNA samples were prepared from either the wild-type (wt; YAS2276) or spb8-2 (YAS2277) strain carrying the GAL1:PGK1NSpG reporter mRNA (pRP611) (29). Yeast cells were grown in galactose minimal medium and then shifted to glucose for the indicated times. PGK1NSpG mRNAs were detected by Northern blot analysis with an end-labeled oligo(C) probe. Positions of the PGK1NSpG mRNA and the SCR1 RNA loading control are indicated.
Similar articles
- Decapping of stabilized, polyadenylated mRNA in yeast pab1 mutants.
Morrissey JP, Deardorff JA, Hebron C, Sachs AB. Morrissey JP, et al. Yeast. 1999 Jun 15;15(8):687-702. doi: 10.1002/(SICI)1097-0061(19990615)15:8<687::AID-YEA412>3.0.CO;2-L. Yeast. 1999. PMID: 10392446 - Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae.
Dunckley T, Tucker M, Parker R. Dunckley T, et al. Genetics. 2001 Jan;157(1):27-37. doi: 10.1093/genetics/157.1.27. Genetics. 2001. PMID: 11139489 Free PMC article. - mRNA decapping activities and their biological roles.
LaGrandeur TE, Parker R. LaGrandeur TE, et al. Biochimie. 1996;78(11-12):1049-55. doi: 10.1016/s0300-9084(97)86729-6. Biochimie. 1996. PMID: 9150884 Review. - An essential component of the decapping enzyme required for normal rates of mRNA turnover.
Beelman CA, Stevens A, Caponigro G, LaGrandeur TE, Hatfield L, Fortner DM, Parker R. Beelman CA, et al. Nature. 1996 Aug 15;382(6592):642-6. doi: 10.1038/382642a0. Nature. 1996. PMID: 8757137 - Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae.
Tucker M, Parker R. Tucker M, et al. Annu Rev Biochem. 2000;69:571-95. doi: 10.1146/annurev.biochem.69.1.571. Annu Rev Biochem. 2000. PMID: 10966469 Review.
Cited by
- Lsm proteins are required for normal processing of pre-tRNAs and their efficient association with La-homologous protein Lhp1p.
Kufel J, Allmang C, Verdone L, Beggs JD, Tollervey D. Kufel J, et al. Mol Cell Biol. 2002 Jul;22(14):5248-56. doi: 10.1128/MCB.22.14.5248-5256.2002. Mol Cell Biol. 2002. PMID: 12077351 Free PMC article. - Deletion of the PAT1 gene affects translation initiation and suppresses a PAB1 gene deletion in yeast.
Wyers F, Minet M, Dufour ME, Vo LT, Lacroute F. Wyers F, et al. Mol Cell Biol. 2000 May;20(10):3538-49. doi: 10.1128/MCB.20.10.3538-3549.2000. Mol Cell Biol. 2000. PMID: 10779343 Free PMC article. - A brief survey of mRNA surveillance.
van Hoof A, Wagner EJ. van Hoof A, et al. Trends Biochem Sci. 2011 Nov;36(11):585-92. doi: 10.1016/j.tibs.2011.07.005. Epub 2011 Sep 6. Trends Biochem Sci. 2011. PMID: 21903397 Free PMC article. Review. - Analysis of mutations in the yeast mRNA decapping enzyme.
Tharun S, Parker R. Tharun S, et al. Genetics. 1999 Apr;151(4):1273-85. doi: 10.1093/genetics/151.4.1273. Genetics. 1999. PMID: 10101156 Free PMC article. - Analysis of P-body assembly in Saccharomyces cerevisiae.
Teixeira D, Parker R. Teixeira D, et al. Mol Biol Cell. 2007 Jun;18(6):2274-87. doi: 10.1091/mbc.e07-03-0199. Epub 2007 Apr 11. Mol Biol Cell. 2007. PMID: 17429074 Free PMC article.
References
- Ares M, Jr, Igel A H. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev. 1990;4:2132–2145. - PubMed
- Beelman C A, Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J Biol Chem. 1994;269:9687–9692. - PubMed
- Beelman C A, Stevens A, Caponigro G, LaGrandeur T E, Hatfield L, Fortner D M, Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. - PubMed
- Broach J R, Strathern J N, Hicks J B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979;8:121–133. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous