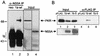Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation - PubMed (original) (raw)
Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation
M Gale Jr et al. Mol Cell Biol. 1998 Sep.
Abstract
The PKR protein kinase is a critical component of the cellular antiviral and antiproliferative responses induced by interferons. Recent evidence indicates that the nonstructural 5A (NS5A) protein of hepatitis C virus (HCV) can repress PKR function in vivo, possibly allowing HCV to escape the antiviral effects of interferon. NS5A presents a unique tool by which to study the molecular mechanisms of PKR regulation in that mutations within a region of NS5A, termed the interferon sensitivity-determining region (ISDR), are associated with sensitivity of HCV to the antiviral effects of interferon. In this study, we investigated the mechanisms of NS5A-mediated PKR regulation and the effect of ISDR mutations on this regulatory process. We observed that the NS5A ISDR, though necessary, was not sufficient for PKR interactions; we found that an additional 26 amino acids (aa) carboxyl to the ISDR were required for NS5A-PKR complex formation. Conversely, we localized NS5A binding to within PKR aa 244 to 296, recently recognized as a PKR dimerization domain. Consistent with this observation, we found that NS5A from interferon-resistant HCV genotype 1b disrupted kinase dimerization in vivo. NS5A-mediated disruption of PKR dimerization resulted in repression of PKR function and inhibition of PKR-mediated eIF-2alpha phosphorylation. Introduction of multiple ISDR mutations abrogated the ability of NS5A to bind to PKR in mammalian cells and to inhibit PKR in a yeast functional assay. These results indicate that mutations within the PKR-binding region of NS5A, including those within the ISDR, can disrupt the NS5A-PKR interaction, possibly rendering HCV sensitive to the antiviral effects of interferon. We propose a model of PKR regulation by NS5A which may have implications for therapeutic strategies against HCV.
Figures
FIG. 1
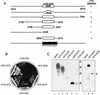
NS5A-binding domain of PKR. (A) Hf7c yeast strains harboring the indicated AD and BD expression constructs were replica printed onto +His (left) and −His (right) media, incubated at 30°C for 4 days, and assayed for growth. Expression of AD-PKR and BD-NS5A 1b-wt constructs was confirmed by immunoblot analysis (not shown). Strains which grew on −His medium were scored positive for a two-hybrid protein interaction. (B) Structural representation of PKR. The positions of the two dsRNA-binding motifs (dsRBM 1 and 2) and the 11 protein kinase catalytic domain conservation regions (roman numerals) (28) are indicated in black. The regions of PKR which mediate interaction with the virus-encoded inhibitors adenovirus VA1 RNA (38), vaccinia virus K3L (16, 25), and HCV NS5A proteins (reference and this study) are underlined.
FIG. 2
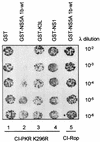
NS5A disrupts PKR dimerization. E. coli AG1688 cells were cotransformed with expression plasmid combinations encoding cI-PKR K296R and GST (column 1), GST-NS5A 1b-wt (column 2), GST-K3L (column 3), or GST-NS1 (column 4), mixed with the indicated dilution of λKH54 phage, and spotted onto plates containing agar medium. Plates were incubated and visually scored for colony formation (dark spots) as described in Materials and Methods. Column 5 contains E. coli harboring cI-Rop and GST-NS5A 1b-wt expression plasmids (control). cI fusion protein dimerization is indicated by colony formation. Shown is a photograph of a plate from a representative experiment.
FIG. 3
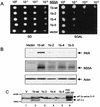
PKR-binding domain of NS5A. (A) Structural representation of BD-NS5A fusion constructs. Deletion mutants were prepared from NS5A 1b-wt, except for the ΔISDR construct, which was prepared from NS5A 1a-wt (27). The ISDR and the PKR-binding region are shown as white and black rectangles, respectively. Terminal amino acid positions are indicated, with numbering based on the prototypic HCV-J polyprotein sequence (36). The PKR interaction of each construct (scored in panel B) is indicated at the right. (B) Yeast two-hybrid assay. Hf7c yeast strains harboring AD-PKR K296R were cotransformed with the indicated BD-NS5A deletion constructs. Strains were propagated on +His medium for 3 days (not shown), after which single colonies were streaked onto −His medium and assayed for growth. Shown is a −His plate incubated for 3 days at 30°C. Growth on −His medium is indicative of a two-hybrid protein interaction. In parallel experiments, we determined that the indicated BD-NS5A constructs did not interact with a construct encoding the GAL4 AD alone (not shown). (C) Immunoblot analysis. Extracts prepared from the yeast strains shown in panel B were separated by SDS-PAGE and subjected to immunoblot analysis using anti-NS5A (lanes 1 to 6) or anti-BD (lanes 7 to 9) monoclonal antibody. Lanes 1 and 7 contain extracts prepared from strains harboring the pGBT9 BD vector (control). Extracts are identified by the construct designation shown above the corresponding lane. BD-NS5A construct 1973-2419 was included as a positive control for the blot shown at the right. Positions of protein standards are indicated in kilodaltons. Arrow points to the protein expressed by the 1973-2208 construct.
FIG. 4
Effects of ISDR mutations on the NS5A-PKR interaction. (A) Yeast two-hybrid assay. Hf7c yeast strains harboring pGAD425 encoding AD-PKR K296R (AD-PKR) or the AD alone (AD-vector) were cotransformed with pGBT9 encoding the BD alone (vector), BD-NS5A 1a-wt, BD-NS5A-ΔISDR, or an isogenic set of BD-NS5A 1b-wt constructs possessing the ISDR sequence shown in Table 3. Strains were replica printed onto +His (left) and −His (right) media and incubated for 3 days at 30°C. Growth on −His medium is indicative of a two-hybrid protein interaction. (B) Immunoblot analysis. Extracts were prepared from the strains shown in panel A and subjected to immunoblot analysis using anti-AD (left) or anti-BD (right) monoclonal antibody. Lanes 1 and 2 show expression of the AD vector (V; control) and AD-PKR (PKR; arrow), respectively. Lanes 3 to 9 show expression of the BD vector (V; lane 3), BD-NS5A-ΔISDR (Δ; lane 4), BD-NS5A 1a-wt (wt; lane 5), BD-NS5A 1b-wt (wt; lane 6), BD-NS5A 1b-2 (2; lane 7), BD-NS5A 1b-4 (4; lane 8), and BD-NS5A 1b-5 (5; lane 9). Arrows at the right indicate positions of the ΔISDR and full-length BD-NS5A constructs. Positions of protein standards are shown in kilodaltons.
FIG. 5
ISDR mutations abolish NS5A function. (A) Yeast growth assay. Cell equivalents of RY1-1 yeast strains harboring the galactose-inducible URA3 expression plasmid pYES-NS5A 1b-wt (1b-wt), pYES-NS5A 1b-2 (1b-2), pYES-NS5A 1b-4 (1b-4), or pYES-NS5A 1b-5 (1b-5) or the pYES control (vector) were serially diluted and spotted onto SD or SGAL medium. Panels show colony formation after 5 days growth at 30°C. (B) Immunoblot analysis of protein extracts prepared from the yeast strains shown in panel A, probed sequentially with anti-PKR, anti-NS5A, and anti-actin (control) monoclonal antibodies. Arrows at the right denote positions of PKR, NS5A, and actin. Each lane represents 50 μg of total protein. (C) eIF-2α phosphorylation. Extracts prepared from the yeast strains shown in panel A were separated by single-dimension IEF and blotted onto a nitrocellulose membrane. Detection of eIF-2α was facilitated by probing the blot with anti-yeast eIF-2α serum. Each lane represents 20 μg of protein prepared from RY1-1 cells harboring pYES (V; lane 1), pYES-NS5A 1b-wt (1b-wt; lane 2), pYES-NS5A 1b-2 (1b-2; lane 3), pYES-NS5A 1b-4 (1b-4; lane 4), pYES-NS5A 1b-5 (1b-5; lane 5), or pYex-PKRΔ295-300 (PKR Δ295-300 [control]; lane 6). Arrows at the right show positions of hypophosphorylated eIF-2α (lower) and hyperphosphorylated eIF-2α, which is phosphorylated by PKR on serine 51. Bars at the left identify the acidic and basic ends of the blot.
FIG. 6
ISDR mutations disrupt the NS5A-PKR association in mammalian cells. Cos-1 cells were cotransfected with cytomegalovirus expression plasmids encoding PKR K296R and NS5A or with PKR K296R and the vector control. Extracts were prepared and mixed with anti-NS5A monoclonal antibody (A) or anti-FLAG resin (B). (A) Anti-NS5A immunocomplexes prepared from extracts harboring PKR K296R with vector control (neo; lane 1) or NS5A 1a-wt (1a-wt; lane 2) and input extract (Input; lanes 3 and 4) were separated by SDS-PAGE and subjected to immunoblot analysis using anti-PKR (lanes 1 to 3) or anti-NS5A (lane 4) monoclonal antibody. Lanes 3 and 4 represent the starting material from the immunoprecipitation (IP) reaction shown in lane 2. The vertical line at left indicates the broad band corresponding to the immunoglobulin (Ig) heavy chain. Positions of protein standards are indicated in kilodaltons. (B) Immunoblot analysis of input protein (lanes 1 to 3) or protein complexes (lanes 4 to 6) recovered by mixing extracts harboring PKR K296R with vector alone (pFLAG; lanes 1 and 4), FLAG-NS5A 1b-wt (1b-wt; lanes 2 and 5), or FLAG-NS5A 1b-5 (1b-5; lanes 3 and 6) with anti-FLAG resin. Blots were probed with a monoclonal antibody specific to human PKR (top) or NS5A (bottom). Arrows point to PKR and NS5A.
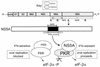
FIG. 7
Role of NS5A in PKR regulation during HCV infection. HCV sensitivity to IFN is determined, at least in part, by the structure of the PKR-binding domain (dark region) within the NS5A cleavage product of the HCV polyprotein. During HCV infection NS5A from wt, IFN-resistant strains of HCV binds PKR, disrupting the critical PKR dimerization process. Resulting PKR monomers are unable to phosphorylate eIF-2α, and thus viral replication proceeds unobstructed (lower right). Mutations within the 66-aa PKR-binding region of NS5A, including the ISDR (indicated by bars), abolish the PKR-regulatory properties of HCV, rendering the virus sensitive to the antiviral actions of IFN. In this case, PKR remains active in a dimeric state and phosphorylates eIF-2α to inhibit mRNA translation and viral replication (lower left).
Similar articles
- Mutations in the interferon-sensitivity determining region of hepatitis C virus and transcriptional activity of the nonstructural region 5A protein.
Fukuma T, Enomoto N, Marumo F, Sato C. Fukuma T, et al. Hepatology. 1998 Oct;28(4):1147-53. doi: 10.1002/hep.510280433. Hepatology. 1998. PMID: 9755255 - Repression of the PKR protein kinase by the hepatitis C virus NS5A protein: a potential mechanism of interferon resistance.
Gale MJ Jr, Korth MJ, Katze MG. Gale MJ Jr, et al. Clin Diagn Virol. 1998 Jul 15;10(2-3):157-62. doi: 10.1016/s0928-0197(98)00034-8. Clin Diagn Virol. 1998. PMID: 9741641 Review. - Antiapoptotic and oncogenic potentials of hepatitis C virus are linked to interferon resistance by viral repression of the PKR protein kinase.
Gale M Jr, Kwieciszewski B, Dossett M, Nakao H, Katze MG. Gale M Jr, et al. J Virol. 1999 Aug;73(8):6506-16. doi: 10.1128/JVI.73.8.6506-6516.1999. J Virol. 1999. PMID: 10400746 Free PMC article. - Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein.
Gale MJ Jr, Korth MJ, Tang NM, Tan SL, Hopkins DA, Dever TE, Polyak SJ, Gretch DR, Katze MG. Gale MJ Jr, et al. Virology. 1997 Apr 14;230(2):217-27. doi: 10.1006/viro.1997.8493. Virology. 1997. PMID: 9143277
Cited by
- Unraveling the dynamics of hepatitis C virus adaptive mutations and their impact on antiviral responses in primary human hepatocytes.
Frericks N, Brown RJP, Reinecke BM, Herrmann M, Brüggemann Y, Todt D, Miskey C, Vondran FWR, Steinmann E, Pietschmann T, Sheldon J. Frericks N, et al. J Virol. 2024 Mar 19;98(3):e0192123. doi: 10.1128/jvi.01921-23. Epub 2024 Feb 6. J Virol. 2024. PMID: 38319104 Free PMC article. - Suppression of Innate Immunity by the Hepatitis C Virus (HCV): Revisiting the Specificity of Host-Virus Interactive Pathways.
Barik S. Barik S. Int J Mol Sci. 2023 Nov 8;24(22):16100. doi: 10.3390/ijms242216100. Int J Mol Sci. 2023. PMID: 38003289 Free PMC article. Review. - Directed natural evolution generates a next-generation oncolytic virus with a high potency and safety profile.
Guo L, Hu C, Liu Y, Chen X, Song D, Shen R, Liu Z, Jia X, Zhang Q, Gao Y, Deng Z, Zuo T, Hu J, Zhu W, Cai J, Yan G, Liang J, Lin Y. Guo L, et al. Nat Commun. 2023 Jun 9;14(1):3410. doi: 10.1038/s41467-023-39156-3. Nat Commun. 2023. PMID: 37296165 Free PMC article. - NS5A domain I antagonises PKR to facilitate the assembly of infectious hepatitis C virus particles.
Chen S, Harris M. Chen S, et al. PLoS Pathog. 2023 Feb 16;19(2):e1010812. doi: 10.1371/journal.ppat.1010812. eCollection 2023 Feb. PLoS Pathog. 2023. PMID: 36795772 Free PMC article. - Hepatitis C Virus Nonstructural Protein 5A Interacts with Immunomodulatory Kinase IKKε to Negatively Regulate Innate Antiviral Immunity.
Kang SM, Park JY, Han HJ, Song BM, Tark D, Choi BS, Hwang SB. Kang SM, et al. Mol Cells. 2022 Oct 31;45(10):702-717. doi: 10.14348/molcells.2022.0018. Epub 2022 Aug 22. Mol Cells. 2022. PMID: 35993162 Free PMC article.
References
- Alter M. Epidemiology of hepatitis C in the west. Semin Liver Dis. 1995;15:5–14. - PubMed
- Barber G N, Jagus R, Meurs E F, Hovanessian A G, Katze M G. Molecular mechanisms responsible for malignant transformation by regulatory and catalytic domain variants of the interferon-induced enzyme RNA-dependent protein kinase. J Biol Chem. 1995;270:17423–17428. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P51 RR000166/RR/NCRR NIH HHS/United States
- AI22646/AI/NIAID NIH HHS/United States
- R01 AI022646/AI/NIAID NIH HHS/United States
- RR00166/RR/NCRR NIH HHS/United States
- AI41629/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases