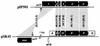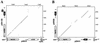Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids - PubMed (original) (raw)
Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids
T Berg et al. J Bacteriol. 1998 Sep.
Abstract
The 46.4-kb nucleotide sequence of pSK41, a prototypical multiresistance plasmid from Staphylococcus aureus, has been determined, representing the first completely sequenced conjugative plasmid from a gram-positive organism. Analysis of the sequence has enabled the identification of the probable replication, maintenance, and transfer functions of the plasmid and has provided insights into the evolution of a clinically significant group of plasmids. The basis of deletions commonly associated with pSK41 family plasmids has been investigated, as has the observed insertion site specificity of Tn552-like beta-lactamase transposons within them. Several of the resistance determinants carried by pSK41-like plasmids were found to be located on up to four smaller cointegrated plasmids. pSK41 and related plasmids appear to represent a consolidation of antimicrobial resistance functions, collected by a preexisting conjugative plasmid via transposon insertion and IS257-mediated cointegrative capture of other plasmids.
Figures
FIG. 1
Maps of the staphylococcal conjugative plasmids, pSK41, pJE1, pGO1, and pUW3626 (3, 9, 11, 34); plasmid sizes are shown on the right. The genetic loci shown are aacA-aphD (Gmr Tmr Kmr), aadD (Nmr), ble (bleomycin resistance), dfrA (trimethoprim resistance), smr (antiseptic and disinfectant resistance), blaZ (penicillin resistance), tra/trs (conjugative transfer functions), and oriT (origin of conjugative transfer). The positions and extents of the cointegrated copy of the plasmid pUB110 (4), the Tn_4001_-IS_257_ hybrid structure (3), and the Tn_552_-like transposon (15) are indicated. Truncated copies of IS_256_ are represented by open boxes, whereas IS_257_ elements are represented as solid boxes containing an arrowhead indicating the direction of transposase transcription and hence the element’s orientation. IS_257_ element designations, following the nomenclature of Leelaporn et al. (23), are shown for pSK41 and pJE1, whereas designations for pGO1 are taken from Morton et al. (34). Where known, the 8 nt of sequence adjacent to each IS_257_ element is indicated; flanking sequences on pGO1 are from Morton et al. (33, 34). _Eco_RI restriction sites (E) are shown.
FIG. 2
Genetic organization of pSK41. Genes are represented by open boxes with names shown within or below and with arrowheads indicating the direction of transcription; smaller open boxes represent interrupted genes. Copies of IS_257_ are denoted by black boxes containing the element’s designation (A through G), whereas inverted copies of truncated IS_256_ elements are indicated by open triangles. Hatched segments indicate integrated small plasmids. RC plasmid replication-initiation genes are suffixed (RC) to differentiate them from the probable theta-mode rep gene of pSK41. Kilobase coordinates are shown below, as are the positions of Eco_RI restriction sites (E) and oriT (vertical arrow). The position in pSK41 corresponding to the insertion site of the Tn_552_-like transposon in pUW3626 is indicated by an asterisk (∗). The position and genetic organization of the additional DNA segment present in pJE1 is also indicated; the tnp genes of the IS_257 elements have been omitted for clarity. The pSK639-like rep gene remnant is suffixed (639) to differentiate it from pSK41 rep. Genetic nomenclature is as described for Fig. 1 and as follows: numbers, size in codons of deduced ORF of unknown function; res, resolvase; oriT, origin of conjugative transfer; nes, oriT nickase; rep/repU, replication initiation; tnp, transposase; pre, recombinase; artA/traA-M, transfer-associated genes; thyE, thymidilate synthetase.
FIG. 3
(A) Phylogenetic analysis of the deduced pSK41 rep product and related proteins. The amino acid sequences were obtained from the following GenBank entries: pSX267, X92404; pCF10, L14285; pPD1, D78016; pAD1, L01794; pLJ1, J04240; pSAK1, Z50862; and IREBF-1, M55290. The unrooted tree was constructed by using the programs PROTDIST and NEIGHBOR (10) from a multiple alignment generated by PILEUP (8). An equivalent tree was obtained with the program PROTPARS (10). (B) Nucleotide sequence alignment of the replication regions of pSK41 and pSX267 (13); identities are indicated by vertical lines and indels are denoted by dots. The nucleotide sequences are numbered on the right. Ribosome binding sites for the rep genes are underlined, and the deduced amino acid sequences are shown above and below the nucleotide sequences, respectively. Directly repeated sequences are indicated by arrowed lines, whereas inverted repeats are shown by half-arrowed lines. Amino acids conserved in all proteins shown in panel A are boxed.
FIG. 4
Relatedness of pSK41 and pIP501 conjugation systems. The genetic organization of the pIP501 conjugation region A (55) and pSK41 nes/oriT and artA-traG of the tra region (11) is illustrated. Genes (arrowed boxes) encoding homologous products are linked by shading; percent amino acid sequence identities and Z scores (in parentheses) are shown. The positions and strands of the oriT nick site of each plasmid are indicated by vertical arrows. A segment of approximately 11 kb between the two regions of pSK41 has been omitted for clarity.
FIG. 5
Dot plot comparisons between segments of pSK41 (x axes) and the small RC plasmids (y axes), pSK108 (A) and pSK89 (B). Dots indicate 20-nt stretches containing at least 17 identities. The genetic organization of the plasmids is illustrated beside the relevant axes. The extents and directions of genes are indicated by arrowed boxes; homologous genes are indicated by equivalent shading. The locations of the SSOA sequences are denoted by open boxes, and the positions of the putative replication nick sites are indicated by an arrow. The locations of the flanking IS_257_ elements in pSK41 are also shown. The segment of pSK108 orf334 not represented in the comparable pSK41 sequence is indicated by hatching. Sequence numbering for pSK89 and pSK108 starts at the first nucleotide their replication-initiation genes, rep and orf334, respectively. The pSK108 (22) and pSK89 (25) sequences are from the GenBank entries U15783 and M37889, respectively.
FIG. 6
Comparison of IS_257_ elements from pSK41, pJE1, and pSK639. Element designations are given on the left, whereas sequence numbering is shown on the right. The pSK639 sequences are taken from GenBank entry U40259. Dots indicate identity to the top sequence, whereas dashes indicate the positions of indels. The terminal inverted repeats of IS_257_ are indicated by arrowed lines. The position of the proposed crossover sites in IS_257_5093B and IS_257_5094E can be localized to the regions between the brackets in these sequences.
Similar articles
- Replication of staphylococcal multiresistance plasmids.
Firth N, Apisiridej S, Berg T, O'Rourke BA, Curnock S, Dyke KG, Skurray RA. Firth N, et al. J Bacteriol. 2000 Apr;182(8):2170-8. doi: 10.1128/JB.182.8.2170-2178.2000. J Bacteriol. 2000. PMID: 10735859 Free PMC article. - Biology of the staphylococcal conjugative multiresistance plasmid pSK41.
Liu MA, Kwong SM, Jensen SO, Brzoska AJ, Firth N. Liu MA, et al. Plasmid. 2013 Jul;70(1):42-51. doi: 10.1016/j.plasmid.2013.02.001. Epub 2013 Feb 13. Plasmid. 2013. PMID: 23415796 Review. - Analysis of a transfer region from the staphylococcal conjugative plasmid pSK41.
Firth N, Ridgway KP, Byrne ME, Fink PD, Johnson L, Paulsen IT, Skurray RA. Firth N, et al. Gene. 1993 Dec 22;136(1-2):13-25. doi: 10.1016/0378-1119(93)90442-6. Gene. 1993. PMID: 8293996 - Processing of Nonconjugative Resistance Plasmids by Conjugation Nicking Enzyme of Staphylococci.
Pollet RM, Ingle JD, Hymes JP, Eakes TC, Eto KY, Kwong SM, Ramsay JP, Firth N, Redinbo MR. Pollet RM, et al. J Bacteriol. 2016 Jan 4;198(6):888-97. doi: 10.1128/JB.00832-15. J Bacteriol. 2016. PMID: 26728193 Free PMC article. - Multiresistant Staphylococcus aureus: genetics and evolution of epidemic Australian strains.
Skurray RA, Rouch DA, Lyon BR, Gillespie MT, Tennent JM, Byrne ME, Messerotti LJ, May JW. Skurray RA, et al. J Antimicrob Chemother. 1988 Apr;21 Suppl C:19-39. doi: 10.1093/jac/21.suppl_c.19. J Antimicrob Chemother. 1988. PMID: 2838448 Review.
Cited by
- Native plasmids of Fusobacterium nucleatum: characterization and use in development of genetic systems.
Haake SK, Yoder SC, Attarian G, Podkaminer K. Haake SK, et al. J Bacteriol. 2000 Feb;182(4):1176-80. doi: 10.1128/JB.182.4.1176-1180.2000. J Bacteriol. 2000. PMID: 10648549 Free PMC article. - The tra region of the conjugative plasmid pIP501 is organized in an operon with the first gene encoding the relaxase.
Kurenbach B, Grothe D, Farías ME, Szewzyk U, Grohmann E. Kurenbach B, et al. J Bacteriol. 2002 Mar;184(6):1801-5. doi: 10.1128/JB.184.6.1801-1805.2002. J Bacteriol. 2002. PMID: 11872736 Free PMC article. - Plasmid content of a vancomycin-resistant Enterococcus faecalis isolate from a patient also colonized by Staphylococcus aureus with a VanA phenotype.
Flannagan SE, Chow JW, Donabedian SM, Brown WJ, Perri MB, Zervos MJ, Ozawa Y, Clewell DB. Flannagan SE, et al. Antimicrob Agents Chemother. 2003 Dec;47(12):3954-9. doi: 10.1128/AAC.47.12.3954-3959.2003. Antimicrob Agents Chemother. 2003. PMID: 14638508 Free PMC article. - Antagonism between aminoglycosides and beta-lactams in a methicillin-resistant Staphylococcus aureus isolate involves induction of an aminoglycoside-modifying enzyme.
Ida T, Okamoto R, Nonoyama M, Irinoda K, Kurazono M, Inoue M. Ida T, et al. Antimicrob Agents Chemother. 2002 May;46(5):1516-21. doi: 10.1128/AAC.46.5.1516-1521.2002. Antimicrob Agents Chemother. 2002. PMID: 11959590 Free PMC article. - Staphylococcal Plasmids, Transposable and Integrative Elements.
Firth N, Jensen SO, Kwong SM, Skurray RA, Ramsay JP. Firth N, et al. Microbiol Spectr. 2018 Nov;6(6):10.1128/microbiolspec.gpp3-0030-2018. doi: 10.1128/microbiolspec.GPP3-0030-2018. Microbiol Spectr. 2018. PMID: 30547857 Free PMC article. Review.
References
- Apisiridej S, Leelaporn A, Scaramuzzi C D, Skurray R A, Firth N. Molecular analysis of a mobilizable theta-mode trimethoprim resistance plasmid from coagulase-negative staphylococci. Plasmid. 1997;38:13–24. - PubMed
- Byrne M E, Gillespie M T, Skurray R A. 4′,4" adenyltransferase activity on conjugative plasmids isolated from Staphylococcus aureus is encoded on an integrated copy of pUB110. Plasmid. 1991;25:70–75. - PubMed
- Claros M G, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources