Pathway choice in glutamate synthesis in Escherichia coli - PubMed (original) (raw)
Pathway choice in glutamate synthesis in Escherichia coli
R B Helling. J Bacteriol. 1998 Sep.
Abstract
Escherichia coli has two primary pathways for glutamate synthesis. The glutamine synthetase-glutamate synthase (GOGAT) pathway is essential for synthesis at low ammonium concentration and for regulation of the glutamine pool. The glutamate dehydrogenase (GDH) pathway is important during glucose-limited growth. It has been hypothesized that GDH is favored when the organism is stressed for energy, because the enzyme does not use ATP as does the GOGAT pathway. The results of competition experiments between the wild-type and a GDH-deficient mutant during glucose-limited growth in the presence of the nonmetabolizable glucose analog alpha-methylglucoside were consistent with the hypothesis. Enzyme measurements showed that levels of the enzymes of the glutamate pathways dropped as the organism passed from unrestricted to glucose-restricted growth. However, other conditions influencing pathway choice had no substantial effect on enzyme levels. Therefore, substrate availability and/or modulation of enzyme activity are likely to be major determinants of pathway choice in glutamate synthesis.
Figures
FIG. 1
Comparison of GS levels during unlimited and glucose-limited growth. (Left) Total (upper) and unadenylylated (lower) enzyme activity. (Right) Average number of active monomers in the holoenzyme, as a function of dilution rate. The maximum specific growth rate is 0.44 h−1.
FIG. 2
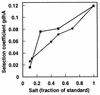
Growth disadvantage of a gdhA mutant during glucose-limited growth as a function of ammonium or phosphate concentration. Ammonium (■) and phosphate (•) concentrations are represented as fractions of those in 1× medium (see Materials and Methods). Data are taken from reference .
FIG. 3
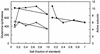
GS during glucose-limited growth as a function of ammonium or phosphate concentration. (Left panel) The uppermost curves (•, ■) show total enzyme activity from RH830 cells, and the lowermost curves (○, □) represent unadenylylated GS activity. (Right panel) Average number of active monomers in the 12-monomer holoenzyme. Ammonium (■, □) and phosphate (•, ○) concentrations are represented as fractions of those in 1× medium (see Materials and Methods).
FIG. 4
GS during unrestricted growth on glucose as a function of ammonium or phosphate concentration. (Left panel) The uppermost curves (•, ■) show total enzyme activity from RH830 and the lowermost curves (○, □) represent unadenylylated GS activity. (Right panel) Average number of active monomers in the holoenzyme. Ammonium (■, □) and phosphate (•, ○) concentrations are represented as fractions of those in 1× medium.
FIG. 5
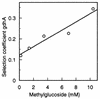
The growth disadvantage of glucose-limited cells lacking GDH as a function of the concentration of the nonmetabolizable glucose analog α-methylglucoside.
FIG. 6
GS during glucose-restricted growth in the presence of the nonmetabolizable glucose analog αMG. (Left panel) Total (upper) and unadenylylated (lower) enzyme activity from strain RH830. (Right panel) average number of active monomers in the holoenzyme.
Similar articles
- Why does Escherichia coli have two primary pathways for synthesis of glutamate?
Helling RB. Helling RB. J Bacteriol. 1994 Aug;176(15):4664-8. doi: 10.1128/jb.176.15.4664-4668.1994. J Bacteriol. 1994. PMID: 7913929 Free PMC article. - Flux balance analysis of ammonia assimilation network in E. coli predicts preferred regulation point.
Wang L, Lai L, Ouyang Q, Tang C. Wang L, et al. PLoS One. 2011 Jan 25;6(1):e16362. doi: 10.1371/journal.pone.0016362. PLoS One. 2011. PMID: 21283535 Free PMC article. - Ammonia assimilation and glutamate formation in the anaerobe Selenomonas ruminantium.
Smith CJ, Hespell RB, Bryant MP. Smith CJ, et al. J Bacteriol. 1980 Feb;141(2):593-602. doi: 10.1128/jb.141.2.593-602.1980. J Bacteriol. 1980. PMID: 6102549 Free PMC article. - Regulation of the assimilation of nitrogen compounds.
Tyler B. Tyler B. Annu Rev Biochem. 1978;47:1127-62. doi: 10.1146/annurev.bi.47.070178.005403. Annu Rev Biochem. 1978. PMID: 28074 Review. No abstract available.
Cited by
- Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50.
Cui L, Murakami H, Kuwahara-Arai K, Hanaki H, Hiramatsu K. Cui L, et al. Antimicrob Agents Chemother. 2000 Sep;44(9):2276-85. doi: 10.1128/AAC.44.9.2276-2285.2000. Antimicrob Agents Chemother. 2000. PMID: 10952568 Free PMC article. - Metabolic regulation of Escherichia coli and its gdhA, glnL, gltB, D mutants under different carbon and nitrogen limitations in the continuous culture.
Kumar R, Shimizu K. Kumar R, et al. Microb Cell Fact. 2010 Jan 27;9:8. doi: 10.1186/1475-2859-9-8. Microb Cell Fact. 2010. PMID: 20105320 Free PMC article. - Broad time-dependent transcriptional activity of metabolic genes of E. coli O104:H4 strain C227/11Φcu in a soil microenvironment at low temperature.
Detert K, Währer J, Nieselt K, Schmidt H. Detert K, et al. Environ Microbiol Rep. 2023 Dec;15(6):582-596. doi: 10.1111/1758-2229.13198. Epub 2023 Aug 29. Environ Microbiol Rep. 2023. PMID: 37644642 Free PMC article. - Transcriptomes of the Extremely Thermoacidophilic Archaeon Metallosphaera sedula Exposed to Metal "Shock" Reveal Generic and Specific Metal Responses.
Wheaton GH, Mukherjee A, Kelly RM. Wheaton GH, et al. Appl Environ Microbiol. 2016 Jul 15;82(15):4613-4627. doi: 10.1128/AEM.01176-16. Print 2016 Aug 1. Appl Environ Microbiol. 2016. PMID: 27208114 Free PMC article. - Achieving optimal growth through product feedback inhibition in metabolism.
Goyal S, Yuan J, Chen T, Rabinowitz JD, Wingreen NS. Goyal S, et al. PLoS Comput Biol. 2010 Jun 3;6(6):e1000802. doi: 10.1371/journal.pcbi.1000802. PLoS Comput Biol. 2010. PMID: 20532205 Free PMC article.
References
- Guest J R. The Leeuwenhoek lecture, 1995. Adaptation to life without oxygen. Philos Trans R Soc Lond B. 1995;350:189–202. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases