Mutations in toxR and toxS that separate transcriptional activation from DNA binding at the cholera toxin gene promoter - PubMed (original) (raw)
Mutations in toxR and toxS that separate transcriptional activation from DNA binding at the cholera toxin gene promoter
J D Pfau et al. J Bacteriol. 1998 Sep.
Abstract
ToxR and ToxS are integral membrane proteins that activate the transcription of virulence genes in Vibrio cholerae. ToxR can be separated into three different domains: an N-terminal cytoplasmic DNA binding domain, a central transmembrane domain, and a C-terminal periplasmic domain. ToxS is thought to enhance ToxR-mediated transcriptional activation through a periplasmic interaction. By P22 challenge phage selection for DNA binding, in combination with a screen for cholera toxin gene transcription, 12 toxR and toxS positive control mutants producing variant ToxR proteins from the toxRS operon that bind to the cholera toxin promoter but that fail to activate transcription were isolated. One mutation in toxR specifies an E82K change in the predicted helix-loop-helix DNA binding domain and destroys ToxR-mediated activation. Seven toxR mutations included frameshifts and stop codons introduced into the periplasmic domain, and six of these mutations appeared to produce proteolytically processed shorter forms of ToxR, suggesting that even short periplasmic deletions alter the folding of ToxR in the periplasm. Deletion of toxS did not alter the steady-state level of ToxR, and ToxR was found to be capable of binding to DNA in the absence of ToxS even though it did not activate transcription. However, the ToxS L33S variant rendered ToxR susceptible to proteolysis, suggesting that the natural function of ToxS is to complex with ToxR. Therefore, certain alterations that map to the ToxR cytoplasmic DNA binding domain, to the periplasmic domain, or to ToxS separate DNA binding activity from activator function. These data support a model where proper assembly or stability of the periplasmic domain of ToxR is enhanced by ToxS. This chaperone-like activity of ToxS may be required for the formation of the transcriptional activation complex but not the ToxR-DNA complex.
Figures
FIG. 1
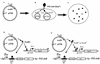
Details of the two-layered selection and screen for positive control mutations in toxRS that retain ToxR-DNA binding activity but that are defective for transcriptional activation (toxR* and toxS* alleles). The system utilizes the challenge phage selection for ToxR-DNA binding in combination with a genetic screen based on loss of ToxRS-dependent activation of a ctx-lacZ promoter fusion. (A to C) Steps utilized for the isolation of mutants; (D and E) molecular basis for the selection and screen. (A) Plasmid pTSK served as the source of toxRS as well as the target for mutagenesis. Plasmid DNA was mutagenized by passage through mutator strains of S. typhimurium (mutD or mutS) or by PCR-mediated region-directed mutagenesis of toxRS. (B) Mutagenized plasmid DNA was introduced into JDP152 carrying a plasmid-borne (pCTX7) ctx-lacZ reporter. Wild-type toxRS activates transcription of the ctx-lacZ fusion, whereas toxRS alleles that are defective for transcriptional activation do not. Pooled bacterial clones were grown to exponential phase in LB medium, infected with challenge phage P22 ctx8, and plated on MacConkey lactose indicator media supplemented with appropriate antibiotics, including kanamycin. Because the decision between lytic and lysogenic development of P22 ctx8 is dependent on ToxR-DNA binding, only toxRS alleles that retain DNA binding activity survive as kanamycin-resistant lysogens. (C) Among the survivors, a subset carried toxR* or toxS* alleles that were defective for transcriptional activation and grew as white-to-pink colonies on MacConkey lactose media. Filled circles symbolize colonies carrying toxR+ and toxS+, and open circles symbolize colonies carrying toxR* or toxS* alleles. Clones carrying the toxR* or toxS* alleles were collected for further analysis. Molecular interactions in the two-layered selection and screen for toxR* or toxS* mutants are shown (D and E). Wild-type ToxRS protein interacts with pCTX7 to activate the transcription of the ctx-lacZ fusion (D), while ToxR simultaneously represses the transcription of the antirepressor gene (ant) carried by the P22 ctx8 challenge phage. The protein encoded by a toxR* or toxS* allele (symbolized by an X in panel E) disrupts the transcriptional activation of ctx-lacZ by ToxRS, but the P22 ctx8 challenge phage is channeled into lysogenic development by ToxR-mediated repression of P22 ant.
FIG. 2
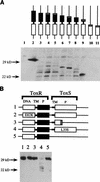
Schematic representation of toxR and toxS alleles that are defective for transcriptional activation but that still retain ToxR-DNA binding activity. The wild-type ToxR and ToxS proteins are shown at the top. The putative ToxR-DNA binding motif (DNA) is shown as an open box, the transmembrane domain (TM) is shown as a dark line, and the periplasmic domain (P) is shown as a filled box. ToxS has a transmembrane domain close to the N terminus. Frameshift peptides are signified by a hatched box. The length of the toxR or toxS coding sequence is shown with the length of the frameshift peptide (if any) in parentheses. Mutations toxS2 and toxS3 carry the change L33S. Allele toxS2 was isolated by region-directed PCR-mediated mutagenesis and carried a silent secondary mutation in the nontranslated region between toxR and toxS. To confirm that the L33S change was solely responsible for the phenotype, toxS3 (L33S) was constructed by site-directed mutagenesis. The Δ_toxS_ and cytoplasmic variant ToxRΔTMHistag constructions are described in Materials and Methods. The ability of each derivative to activate the transcription of ctx-lacZ is shown. A + indicates >90% activity, and a − indicates activity below 25% of that of the wild type. DNA binding activity as measured by the challenge phage assay with phage P22 ctx8 is indicated. A + signifies full DNA binding activity (fraction survival of >5 × 10−3), and a − indicates fraction survival of <1 × 10−4.
FIG. 3
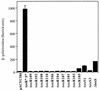
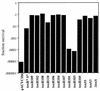
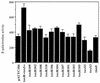
Activation of ctx-lacZ in E. coli by toxR or toxS positive control alleles. Plasmid pTSK or pTSK variants were the source of toxRS expression in these assays. Wild-type ToxRS activates transcription from the V. cholerae O395 ctxAB promoter carried on a lysogenic lambda phage, λ(ctx-lacZ)7, in E. coli JDP169. β-Galactosidase activity is shown in Barrick units for each mutant allele. Plasmid JDP169/pACYC184 is included as a _toxRS_-negative control.
FIG. 4
Challenge phage assay results showing ToxR-DNA binding activity produced by toxR or toxS positive control alleles. Salmonella strain MS1868 carrying pTSK (toxR+ toxS+) or each mutant allele was grown to middle-log phase in LB media at 37°C and infected with P22 ctx8 carrying the ToxR binding site of the ctx promoter from V. cholerae 569B. Challenge phage assays are described in Materials and Methods. ToxR acts as the repressor of P22 lytic development in these assays. Fraction survival is the number of cells surviving infection divided by the number of input cells. Values were averaged from the results of three independent experiments and differed by less than twofold.
FIG. 5
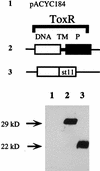
Immunoblot analysis of the steady-state level of ToxR protein produced by pTSK derivatives expressing toxR and toxS positive control alleles. (A) Immunoblot analysis of ToxR truncations produced in vivo in S. typhimurium MS1868. Whole-cell extracts were prepared for each mutant, and proteins were separated on a denaturing SDS-polyacrylamide gel. Cells were grown under conditions identical to those of the challenge phage assays. The proteins were transferred to nitrocellulose, and an anti-ToxR antibody was used to detect cross-reactive species. Schematic representations of each variant ToxR protein are shown above each lane (Fig. 2). Lanes: 1, toxR toxS negative (pACYC184); 2, toxR+ toxS+ (pTSK); 3, toxR109 (L280Oc); 4, toxR102 (E275fs); 5, toxR108 (P271fs); 6, toxR110 (G247Op); 7, toxR106 (K239fs); 8, toxR104 (W229Am); 9, toxR107 (N200fs); 10, toxR101 (M98fs); 11, toxR103 (S93fs). (B) Immunoblot analysis of toxR* or toxS* alleles that carry the full-length toxR coding sequence was performed as described for panel A. Lanes: 1, toxR+ toxS+ (MS1868/pTSK); 2, toxR105 (E82K); 3, toxS1 (E37fs); 4, toxS3 (L33S); 5, Δ_toxS_. See the legend to Fig. 2 for an explanation of symbols and abbreviations.
FIG. 6
Effect of toxR or toxS positive control mutants on the transcription of (ctx-lacZ) fusion strains in the presence of a second plasmid source of toxR+. To test if any of the toxR and toxS mutations are dominant to the activity of toxR+ carried by pVM7, each pTSK mutant toxRS operon was introduced into JDP169/pVM7 [Η(ctx-lacZ) toxR+]. β-Galactosidase activity is shown. JDP169/pACYC184/pBR322 bearing DNA encoding no activators produces 13 Barrick U of β-galactosidase activity (data not shown).
FIG. 7
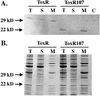
Subcellular localization of ToxR and periplasmic truncation derivative ToxR107. S. typhimurium extracts were fractionated and subjected to immunoblot analysis with an anti-ToxR antibody as described in Materials and Methods. ToxR, MS1868/pTSK (toxR+S+); ToxR107, MS1868/pTSK (toxR107). Lanes: T, total protein; S, soluble protein; M, membrane fraction; C, total protein fraction from the control (MS1868/pACYC184). (A) Immunoblot analysis; (B) Coomassie brilliant blue staining of gel.
FIG. 8
Results of immunoblot assay showing the steady-state level of ToxR and ToxRΔTMHistag under overexpressing conditions. MS1868/F′ _lacI_q/pTacterm, MS1868/F′ _lacI_q/pJAM3 (toxR+), and MS1868/F′ _lacI_q/pJAM3_toxR_ΔHistag were grown and induced with IPTG as described in Materials and Methods for challenge phage assays. Whole-cell extracts were resolved on an SDS-polyacrylamide gel and transferred to nitrocellulose. Membranes were probed with an anti-ToxR antibody. Schematic representations of ToxR and ToxRΔTMHistag are described in the legend to Fig. 2. Lanes: 1, vector control; 2, toxR+; 3, _toxR_ΔTMHistag.
Similar articles
- Host stimuli and operator binding sites controlling protein interactions between virulence master regulator ToxR and ToxS in Vibrio cholerae.
Lembke M, Höfler T, Walter AN, Tutz S, Fengler V, Schild S, Reidl J. Lembke M, et al. Mol Microbiol. 2020 Aug;114(2):262-278. doi: 10.1111/mmi.14510. Epub 2020 Apr 19. Mol Microbiol. 2020. PMID: 32251547 Free PMC article. - Membrane localization of the ToxR winged-helix domain is required for TcpP-mediated virulence gene activation in Vibrio cholerae.
Crawford JA, Krukonis ES, DiRita VJ. Crawford JA, et al. Mol Microbiol. 2003 Mar;47(5):1459-73. doi: 10.1046/j.1365-2958.2003.03398.x. Mol Microbiol. 2003. PMID: 12603748 - Co-ordinate expression of virulence genes by ToxR in Vibrio cholerae.
DiRita VJ. DiRita VJ. Mol Microbiol. 1992 Feb;6(4):451-8. doi: 10.1111/j.1365-2958.1992.tb01489.x. Mol Microbiol. 1992. PMID: 1560773 Review. - Expression of Vibrio cholerae virulence genes in response to environmental signals.
Peterson KM. Peterson KM. Curr Issues Intest Microbiol. 2002 Sep;3(2):29-38. Curr Issues Intest Microbiol. 2002. PMID: 12400636 Review.
Cited by
- Vibrio cholerae H-NS silences virulence gene expression at multiple steps in the ToxR regulatory cascade.
Nye MB, Pfau JD, Skorupski K, Taylor RK. Nye MB, et al. J Bacteriol. 2000 Aug;182(15):4295-303. doi: 10.1128/JB.182.15.4295-4303.2000. J Bacteriol. 2000. PMID: 10894740 Free PMC article. - Proteolysis of virulence regulator ToxR is associated with entry of Vibrio cholerae into a dormant state.
Almagro-Moreno S, Kim TK, Skorupski K, Taylor RK. Almagro-Moreno S, et al. PLoS Genet. 2015 Apr 7;11(4):e1005145. doi: 10.1371/journal.pgen.1005145. eCollection 2015 Apr. PLoS Genet. 2015. PMID: 25849031 Free PMC article. - TcpH influences virulence gene expression in Vibrio cholerae by inhibiting degradation of the transcription activator TcpP.
Beck NA, Krukonis ES, DiRita VJ. Beck NA, et al. J Bacteriol. 2004 Dec;186(24):8309-16. doi: 10.1128/JB.186.24.8309-8316.2004. J Bacteriol. 2004. PMID: 15576780 Free PMC article. - Membrane topology and DNA-binding ability of the Streptococcal CpsA protein.
Hanson BR, Lowe BA, Neely MN. Hanson BR, et al. J Bacteriol. 2011 Jan;193(2):411-20. doi: 10.1128/JB.01098-10. Epub 2010 Nov 19. J Bacteriol. 2011. PMID: 21097630 Free PMC article. - Proteolysis of ToxR is controlled by cysteine-thiol redox state and bile salts in Vibrio cholerae.
Lembke M, Pennetzdorfer N, Tutz S, Koller M, Vorkapic D, Zhu J, Schild S, Reidl J. Lembke M, et al. Mol Microbiol. 2018 Dec;110(5):796-810. doi: 10.1111/mmi.14125. Epub 2018 Oct 25. Mol Microbiol. 2018. PMID: 30218472 Free PMC article.
References
- Amann E, Brosius J, Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983;25:167–178. - PubMed
- Arvidson D N, Youderian P, Schneider T D, Stormo G D. Automated kinetic assay of beta-galactosidase activity. BioTechniques. 1991;11:733–738. - PubMed
- Bianco P R, Weinstock G M. Automated determination of beta-galactosidase specific activity. BioTechniques. 1994;17:974–980. - PubMed
- Bolivar F, Rodriguez R L, Betach M C, Boyer H W. Construction and characterization of new cloning vehicles. I. Ampicillin-resistant derivatives of the plasmid pMB9. Gene. 1977;2:75–93. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases