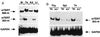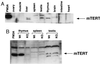Expression of mouse telomerase catalytic subunit in embryos and adult tissues - PubMed (original) (raw)
Expression of mouse telomerase catalytic subunit in embryos and adult tissues
L Martín-Rivera et al. Proc Natl Acad Sci U S A. 1998.
Abstract
Telomerase is a ribonucleoprotein complex that elongates telomeres, allowing the stable maintenance of chromosomes during multiple cell divisions. Here, we describe the isolation and characterization of the catalytic subunit of mouse telomerase, mTERT (mouse telomerase reverse transcriptase), an essential protein component of the telomerase complex. During embryonic development, mTERT mRNA is abundantly expressed in the whole embryo, especially in regions of intense proliferation. We found that the mTERT mRNA expression in both embryonic and adult tissues is independent of the essential RNA component of telomerase, mTR, and therefore, of the formation of active telomerase complexes. mTERT protein is present exclusively in tissues with telomerase activity, such as testis, spleen, and thymus. mTERT protein is barely detectable in the thymus of mTR-/- mice, suggesting that mTERT protein stability in this tissue may depend on the actual assembly of active telomerase complexes. Finally, we found that mouse and human telomerase catalytic subunit is located in the cell nucleus, and its localization is not regulated during cell cycle progression.
Figures
Figure 1
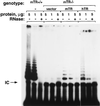
Rescue of telomerase activity in an mTR−/− cell line with the human telomerase RNA. A plasmid with the hTR gene under a constitutive promoter, or with 5 kb of mouse genomic DNA that contained the mTR gene and upstream sequences (15), or an empty vector (Bluescript) were transfected into a mTR−/− cell line (KO-3 at passage 23). Forty-eight hours after transfection, S-100 extracts were prepared and assayed for telomerase activity. As a control of the TRAP assay, telomerase activity was detected in wild-type (mTR+/+) cells. All the extracts were pretreated (+) or not (−) with RNase before the telomerase assay. The protein concentration in the PCR step of the TRAP assay is given in μg/μl. The arrow indicates the position of the internal control (IC) for PCR efficiency.
Figure 2
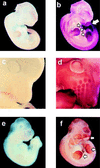
Expression of mTERT mRNA in mouse embryos. (a and b) Eleven-day wild-type embryos hybridized with sense (a) or antisense (b) mTERT-derived riboprobes. b shows generalized expression of mTERT. Regions of particularly high levels of mTERT mRNA are indicated with arrows. (c and d) Detail of the head of a 12-day wild-type embryo hybridized with sense (c) or antisense (d) mTERT-derived riboprobes. d shows high levels of mTERT mRNA in the developing hair follicles of the nares. (e and f) Ten-and-one-half-day mTR−/− embryos hybridized with sense (e) or antisense (f) mTERT-derived riboprobes. The pattern and intensity of the mTERT mRNA signal are similar in b and f.
Figure 3
Expression of mTERT mRNA in adult tissues. Total RNA isolated from the indicated wild-type or mTR−/− mouse tissues was reverse transcribed and then subjected to PCR amplification with two different sets of primers: 998-nt mTERT PCR product was obtained with Race1 and 55/1 primers and a 363-nt product was obtained when clone2a and clone2b primers were used. (A) RT-PCR from four tissues derived from 2-month-old wild-type mice. (B) RT-PCR from three tissues derived from wild-type (wt) or mTR−/− (KO) littermates. Arrows indicate the RT-PCR fragments obtained with primers against mTERT or GAPDH genes. Br, brain; Te, testis; Kd, kidney; Li, liver; Spl, spleen; Thy, thymus.
Figure 4
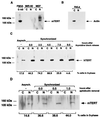
. Characterization of K-370 antibodies against mouse and human telomerase catalytic subunits. Western blot analysis of total cell lysates from the indicated mouse (MEF, FM3A) or human (IMR-90, 293) cells using K-370 antibodies in the absence of competing peptides, or in the presence of the antigenic peptide PEPT-1 or the unrelated peptide PEPT-2. FM3A is a mouse immortal cell line; IMR-90 cells are nonimmortalized human diploid fibroblasts; and 293 cells are adenovirus-transformed human fibroblasts. MEFs had undetectable mTERT protein in this assay and IMR-90 cells are negative for telomerase activity, whereas FM3A, MEFs, and 293 have detectable telomerase activity.
Figure 5
Expression of mTERT protein in different murine tissues. (A) Western blot analysis of S-100 extracts from the indicated tissues, using K-370 antibodies. Tissues were obtained from wild-type mice. One hundred micrograms of total protein was loaded per lane. FM3A is a mouse cell line expressing high levels of mTERT (see Fig. 4). The distortion in the lane corresponding to liver is probably because of the presence of an abundant protein in the relevant region of the gel. Band specificity was confirmed by preincubation of the K-370 antibodies with the antigenic peptide (not shown). (B) Western blot analysis of S-100 extracts of thymus, spleen, and testis obtained from wild-type and mTR−/− littermates, and using K-370 antibodies. One hundred micrograms of total protein was loaded per lane, except in the case of the FM3A extract, of which only 20 μg was loaded. The specificity of the mTERT band was determined by preincubation of the K-370 antibody with the antigenic peptide (not shown).
Figure 6
Subcellular location of human telomerase catalytic subunit. IMR-90 (a and b) and HeLa (c, d, e, and f) cells were simultaneously stained with Hoechst 33258 to visualize nuclei (a, c, and e) and with anti-hTERT antibodies K-370 (b, d, and f). IMR90 are nonimmortal human diploid fibroblasts that have no detectable telomerase activity. HeLa are human papillomavirus-transformed human epithelial cells that have telomerase activity. Note the punctuated hTERT expression pattern in the nucleus. (Objective was ×100.)
Figure 7
Cell cycle regulation of TERT protein. (A) Fifty micrograms of either nuclear (N) or cytoplasmic (C) extracts from asynchronous populations of IMR-90 and MEFs were analyzed by Western blotting using K-370 antibodies. mTERT was detected exclusively in the nucleus of the MEFs. (B) Fifty micrograms of either nuclear (N) or cytoplasmic (C) extracts from asynchronous HeLa cells were analyzed by Western blotting using anti-actin antibodies. The cytoplasmic fraction showed the actin band. (C) Western blot analysis, using K-370 antibodies, of nuclear (N) and cytoplasmic (C) extracts from asynchronous or synchronized HeLa cells at the indicated times after the thymidine block release. Fifty micrograms of total protein was loaded per lane. The percentage of cells in S phase at the different points analyzed is also indicated. (D) Western blot analysis, using K-370 antibodies, of nuclear (N) and cytoplasmic (C) extracts from asynchronous or synchronized MEFs at the indicated times after the aphidicolin block release. One hundred micrograms of total protein was loaded per lane. The percentage of cells in S phase at the indicated different points analyzed is also shown.
Similar articles
- Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation.
Greenberg RA, Allsopp RC, Chin L, Morin GB, DePinho RA. Greenberg RA, et al. Oncogene. 1998 Apr 2;16(13):1723-30. doi: 10.1038/sj.onc.1201933. Oncogene. 1998. PMID: 9582020 - Cloning of rat telomerase catalytic subunit functional domains, reconstitution of telomerase activity and enzymatic profile of pig and chicken tissues.
Wong SC, Ong LL, Er CP, Gao S, Yu H, So JB. Wong SC, et al. Life Sci. 2003 Oct 10;73(21):2749-60. doi: 10.1016/s0024-3205(03)00670-2. Life Sci. 2003. PMID: 13679242 - Presence of telomeric G-strand tails in the telomerase catalytic subunit TERT knockout mice.
Yuan X, Ishibashi S, Hatakeyama S, Saito M, Nakayama J, Nikaido R, Haruyama T, Watanabe Y, Iwata H, Iida M, Sugimura H, Yamada N, Ishikawa F. Yuan X, et al. Genes Cells. 1999 Oct;4(10):563-72. doi: 10.1046/j.1365-2443.1999.00284.x. Genes Cells. 1999. PMID: 10583505 - Mammalian telomerase: catalytic subunit and knockout mice.
Kipling D. Kipling D. Hum Mol Genet. 1997 Nov;6(12):1999-2004. doi: 10.1093/hmg/6.12.1999. Hum Mol Genet. 1997. PMID: 9328462 Review. - Human Specific Regulation of the Telomerase Reverse Transcriptase Gene.
Zhang F, Cheng D, Wang S, Zhu J. Zhang F, et al. Genes (Basel). 2016 Jun 28;7(7):30. doi: 10.3390/genes7070030. Genes (Basel). 2016. PMID: 27367732 Free PMC article. Review.
Cited by
- Novel telomerase-increasing compound in mouse brain delays the onset of amyotrophic lateral sclerosis.
Eitan E, Tichon A, Gazit A, Gitler D, Slavin S, Priel E. Eitan E, et al. EMBO Mol Med. 2012 Apr;4(4):313-29. doi: 10.1002/emmm.201200212. Epub 2012 Feb 20. EMBO Mol Med. 2012. PMID: 22351600 Free PMC article. - Human telomerase RNA-protein interactions.
Bachand F, Triki I, Autexier C. Bachand F, et al. Nucleic Acids Res. 2001 Aug 15;29(16):3385-93. doi: 10.1093/nar/29.16.3385. Nucleic Acids Res. 2001. PMID: 11504876 Free PMC article. - Telomerase reverse transcriptase promotes cardiac muscle cell proliferation, hypertrophy, and survival.
Oh H, Taffet GE, Youker KA, Entman ML, Overbeek PA, Michael LH, Schneider MD. Oh H, et al. Proc Natl Acad Sci U S A. 2001 Aug 28;98(18):10308-13. doi: 10.1073/pnas.191169098. Epub 2001 Aug 21. Proc Natl Acad Sci U S A. 2001. PMID: 11517337 Free PMC article. - Distinct dosage requirements for the maintenance of long and short telomeres in mTert heterozygous mice.
Erdmann N, Liu Y, Harrington L. Erdmann N, et al. Proc Natl Acad Sci U S A. 2004 Apr 20;101(16):6080-5. doi: 10.1073/pnas.0401580101. Epub 2004 Apr 12. Proc Natl Acad Sci U S A. 2004. PMID: 15079066 Free PMC article. - Impaired germinal center reaction in mice with short telomeres.
Herrera E, Martínez-A C, Blasco MA. Herrera E, et al. EMBO J. 2000 Feb 1;19(3):472-81. doi: 10.1093/emboj/19.3.472. EMBO J. 2000. PMID: 10654945 Free PMC article.
References
- Greider C W. Annu Rev Biochem. 1996;65:337–365. - PubMed
- Blasco M A, Lee H-W, Hande P, Samper E, Lansdorp P, DePinho R, Greider C W. Cell. 1997;91:25–34. - PubMed
- Blackburn E H. Nature (London) 1991;350:569–573. - PubMed
- Harley C B, Futcher A B, Greider C W. Nature (London) 1990;345:458–460. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases