A knob-associated tandem repeat in maize capable of forming fold-back DNA segments: are chromosome knobs megatransposons? - PubMed (original) (raw)
A knob-associated tandem repeat in maize capable of forming fold-back DNA segments: are chromosome knobs megatransposons?
E V Ananiev et al. Proc Natl Acad Sci U S A. 1998.
Abstract
A class of tandemly repeated DNA sequences (TR-1) of 350-bp unit length was isolated from the knob DNA of chromosome 9 of Zea mays L. Comparative fluorescence in situ hybridization revealed that TR-1 elements are also present in cytologically detectable knobs on other maize chromosomes in different proportions relative to the previously described 180-bp repeats. At least one knob on chromosome 4 is composed predominantly of the TR-1 repeat. In addition, several small clusters of the TR-1 and 180-bp repeats have been found in different chromosomes, some not located in obvious knob heterochromatin. Variation in restriction fragment fingerprints and copy number of the TR-1 elements was found among maize lines and among maize chromosomes. TR-1 tandem arrays up to 70 kilobases in length can be interspersed with stretches of 180-bp tandem repeat arrays. DNA sequence analysis and restriction mapping of one particular stretch of tandemly arranged TR-1 units indicate that these elements may be organized in the form of fold-back DNA segments. The TR-1 repeat shares two short segments of homology with the 180-bp repeat. The longest of these segments (31 bp; 64% identity) corresponds to the conserved region among 180-bp repeats. The polymorphism and complex structure of knob DNA suggest that, similar to the fold-back DNA-containing giant transposons in Drosophila, maize knob DNA may have some properties of transposable elements.
Figures
Figure 1
Nucleotide sequence of a TR-1 element associated with the knob region of maize chromosome 9. The DNA segment between nucleotides 26 and 56 in the TR-1 element shares 64% homology with the conserved segment between nucleotides 122 and 157 of a 180-bp repeat.
Figure 2
A molecular map of the knob DNA segment with 180-bp and TR-1 tandem repeats from cosmid 9. The FB-1 element is composed of two inverted arrays of TR-1 elements (indicated by two black arrows) and is flanked by 180-bp repeats. Dashed boxes with clear arrows represent blocks of tandem arrays of TR-1 elements, and clear boxes with black short arrows represent blocks of tandem arrays of 180-bp repeats with arrowheads indicating individual repeats and their orientation. E, _Eco_RI site; N, _Nde_I site.
Figure 3
The integration sites of TR-1 elements into 180-bp repeats. All junctures in TR-1 as well as in 180-bp (bold) repeats are different. In three cases, A, C, and D, there are short stretches of unrelated DNA sequences (underlined) that separate the TR-1 from 180-bp repeats.
Figure 4
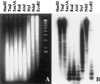
Hybridization of the TR-1 element to a blot panel of maize genomic DNA (Seneca 60) digested with different restriction enzymes. A typical ladder-like pattern of bands hybridizing to TR-1 is seen in lanes with DNA cut by _Eco_RI, _Alu_I, _Rsa_I, and _Sau_3A, the restriction enzymes that have a recognition site within TR-1. DNA samples cut with _Hae_III, _Taq_I, and _Nde_I, which have no recognition sites within TR-1, reveal maximum hybridization signal with DNA fragments larger than 10–20 kb. _Sty_I produced an intermediate ladder-like pattern of hybridization.
Figure 5
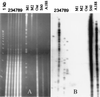
Hybridization of the TR-1 element to a DNA blot panel of oat–maize chromosome addition lines revealed a high level of polymorphism in copy number and restriction DNA fragment pattern among maize chromosomes and also between two maize lines, Seneca 60 and A188. Plant genomic DNA was cut by _Hae_III restriction enzyme. 1kb, molecular weight 1-kb marker ladder; 2, 3, 4, 7, 8, and 9, oat–maize addition lines with corresponding maize chromosomes; M1, mixture of λ DNA cut with _Hin_dIII and _Pst_I; M2, λ monomer (50 kb and dimer 100 kb); Oat, oat DNA from parental line Starter 1; S60 and A188, DNA of two maize lines (Seneca 60 and A188).
Figure 6
In situ hybridization of the TR-1 and 180-bp repeats to pachytene chromosomes of maize (Seneca 60). (A) The overall view of pachytene chromosomes stained with 4′,6-diamino-2-phenylindole (DAPI), the 180-bp repeat (green), and the TR-1 repeat (red) fluorescent images. The 180-bp repeats reveal strong hybridization signal to large knobs on chromosomes 5, 6, and 9, and the TR-1 elements reveal strong hybridization to the large knob on chromosome 4. Several additional small clusters of 180-bp repeats and TR-1 elements may be found in different sites on the chromosomes. (B) Distribution of hybridization signals over chromosomes 4 and 5. Overall view of 4′,6-diamino-2-phenylindole-stained chromosomes 4 and 5 (DAPI), the 180-bp repeat (green), and the TR-1 element (red) fluorescent images. The 180-bp repeat is detected in the large knob of chromosome 5 but not in the large knob of chromosome 4. However, there is a small cluster of 180-bp repeats at the terminus of chromosome 4. The TR-1 repeat is detected in both large knobs and in the small cluster at the terminus of chromosome 4. (C) Distribution of hybridization signals over chromosome 6. The overall view of 4′,6-diamino-2-phenylindole-stained chromosome 6 (DAPI), the 180-bp repeat (green), and the TR-1 element (red) fluorescent images. The 180-bp repeat and TR-1 element form two clusters: a big one in the knob at the terminus of the short arm and a small one probably in the first chromomere of the satellite of chromosome 6.
Similar articles
- Complex structure of knobs and centromeric regions in maize chromosomes.
Ananiev EV, Phillips RL, Rines HW. Ananiev EV, et al. Tsitol Genet. 2000 Mar-Apr;34(2):11-5. Tsitol Genet. 2000. PMID: 10857197 Review. - Complex structure of knob DNA on maize chromosome 9. Retrotransposon invasion into heterochromatin.
Ananiev EV, Phillips RL, Rines HW. Ananiev EV, et al. Genetics. 1998 Aug;149(4):2025-37. doi: 10.1093/genetics/149.4.2025. Genetics. 1998. PMID: 9691055 Free PMC article. - Molecular characterization of a family of tandemly repeated DNA sequences, TR-1, in heterochromatic knobs of maize and its relatives.
Hsu FC, Wang CJ, Chen CM, Hu HY, Chen CC. Hsu FC, et al. Genetics. 2003 Jul;164(3):1087-97. doi: 10.1093/genetics/164.3.1087. Genetics. 2003. PMID: 12871917 Free PMC article. - Fluorescence in situ hybridization analysis reveals multiple loci of knob-associated DNA elements in one-knob and knobless maize lines.
Adawy SS, Stupar RM, Jiang J. Adawy SS, et al. J Histochem Cytochem. 2004 Aug;52(8):1113-6. doi: 10.1369/jhc.4B6335.2004. J Histochem Cytochem. 2004. PMID: 15258188 - Mixed knobs in corn cobs.
Lamelza P, Lampson MA. Lamelza P, et al. Genes Dev. 2020 Sep 1;34(17-18):1110-1112. doi: 10.1101/gad.343350.120. Genes Dev. 2020. PMID: 32873577 Free PMC article. Review.
Cited by
- Intragenomic conflict between the two major knob repeats of maize.
Kanizay LB, Albert PS, Birchler JA, Dawe RK. Kanizay LB, et al. Genetics. 2013 May;194(1):81-9. doi: 10.1534/genetics.112.148882. Epub 2013 Mar 2. Genetics. 2013. PMID: 23457233 Free PMC article. - Comprehending the dynamism of B chromosomes in their journey towards becoming unselfish.
Rajpal VR, Sharma S, Sehgal D, Sharma P, Wadhwa N, Dhakate P, Chandra A, Thakur RK, Deb S, Rama Rao S, Mir BA, Raina SN. Rajpal VR, et al. Front Cell Dev Biol. 2023 Jan 4;10:1072716. doi: 10.3389/fcell.2022.1072716. eCollection 2022. Front Cell Dev Biol. 2023. PMID: 36684438 Free PMC article. Review. - High-resolution crossover maps for each bivalent of Zea mays using recombination nodules.
Anderson LK, Doyle GG, Brigham B, Carter J, Hooker KD, Lai A, Rice M, Stack SM. Anderson LK, et al. Genetics. 2003 Oct;165(2):849-65. doi: 10.1093/genetics/165.2.849. Genetics. 2003. PMID: 14573493 Free PMC article. - Genomic instability within centromeres of interspecific marsupial hybrids.
Metcalfe CJ, Bulazel KV, Ferreri GC, Schroeder-Reiter E, Wanner G, Rens W, Obergfell C, Eldridge MD, O'Neill RJ. Metcalfe CJ, et al. Genetics. 2007 Dec;177(4):2507-17. doi: 10.1534/genetics.107.082313. Genetics. 2007. PMID: 18073443 Free PMC article. - Transposons and genome evolution in plants.
Fedoroff N. Fedoroff N. Proc Natl Acad Sci U S A. 2000 Jun 20;97(13):7002-7. doi: 10.1073/pnas.97.13.7002. Proc Natl Acad Sci U S A. 2000. PMID: 10860963 Free PMC article. Review.
References
- McClintock B. Science. 1929;69:629. - PubMed
- Reeves R G. Genetica (The Hague) 1944;29:141–147. - PubMed
- Longley A A. Bot Rev. 1952;18:399–412.
- McClintock B, Kato Y, Bluemenshein A. Chromosome Constitution of Races of Maize. Chapingo, Mexico: Colegio de Postgraduados; 1981.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources