SCAR, a WASP-related protein, isolated as a suppressor of receptor defects in late Dictyostelium development - PubMed (original) (raw)
SCAR, a WASP-related protein, isolated as a suppressor of receptor defects in late Dictyostelium development
J E Bear et al. J Cell Biol. 1998.
Abstract
G protein-coupled receptors trigger the reorganization of the actin cytoskeleton in many cell types, but the steps in this signal transduction cascade are poorly understood. During Dictyostelium development, extracellular cAMP functions as a chemoattractant and morphogenetic signal that is transduced via a family of G protein-coupled receptors, the cARs. In a strain where the cAR2 receptor gene is disrupted by homologous recombination, the developmental program arrests before tip formation. In a genetic screen for suppressors of this phenotype, a gene encoding a protein related to the Wiskott-Aldrich Syndrome protein was discovered. Loss of this protein, which we call SCAR (suppressor of cAR), restores tip formation and most later development to cAR2(-) strains, and causes a multiple-tip phenotype in a cAR2(+) strain as well as leading to the production of extremely small cells in suspension culture. SCAR-cells have reduced levels of F-actin staining during vegetative growth, and abnormal cell morphology and actin distribution during chemotaxis. Uncharacterized homologues of SCAR have also been identified in humans, mouse, Caenorhabditis elegans, and Drosophila. These data suggest that SCAR may be a conserved negative regulator of G protein-coupled signaling, and that it plays an important role in regulating the actin cytoskeleton.
Figures
Figure 2
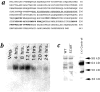
SCAR sequence, developmental Northern blot analysis, and protein expression. (a) Predicted amino acid sequence of SCAR protein derived from cDNA sequence.(∇) Site of plasmid insertion. Underlining indicates highly charged helical region, and residues in bold show polyproline repeats. (b) Developmental Northern blot analysis. At the indicated time points, poly(A)+-selected RNA was prepared from HPS400 cells developed on filter pads, subjected to electrophoresis, blotted, and probed with a 32P-labeled SCAR cDNA probe. The SCAR transcript runs as a single band at 1.8 kb. (c) SCAR protein expression. Whole-cell protein extracts were prepared from HPS400/ SCAR+ and SCAR− cells developed on filters for 17 h, subjected to SDS-PAGE, transferred, and probed with an affinity-purified SCAR polyclonal antisera. This antisera recognizes several bands, including a 60-kD band present only in SCAR+ lysates. (+) Control is a lysate of bacterial cells induced to express a recombinant SCAR fusion protein. This fusion protein includes extra sequences for purification purposes, and runs slightly higher than the endogenous SCAR protein.
Figure 1
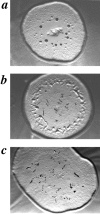
SCAR gene disruption partially rescues carB null morphological block. Developmental phenotypes of carB null strain KO8-5 (a), the original REMI mutant, 9A/O7 (b), and carB null/ SCAR−(c), grown on agar spread with Klebsiella aerogenes.
Figure 3
Developmental phenotypes of HPS400/SCAR+ (a) and HPS400/SCAR− (b) on DB-agar plates at 13 h of development.
Figure 4
Sequence alignments. (a) Sequence alignments of SHD, WH2, and acidic domains for HsSCAR1 (KIAA 0269, NCBI accession no. 1665805), CeSCAR (R06C1.b, NCBI accession no. 1628078), MmSCAR (NCBI accession no. AA035899), DmSCAR (NCBI accession no. AA567420), HsSCAR2 (NCBI accession no. AA535513), HsSCAR3 (NCBI accession no. AA445914), WASP (hWASP, NCBI accession no. 1722836), and Verprolin (NCBI accession no. 755821). Alignments were produced with the LaserGene program (DNASTAR Inc., Madison, WI). Identical residues are outlined in black, and highly conserved residues are outlined in gray. Asterisks show residues that are invariant in all sequences at that position. (b) Domain structure of proteins containing ABM-2 polyproline repeats (Poly-proline). WH2 indicates WASP homology 2 domain; PH indicates Plekstrin homology domain; GBD (also known as a CRIB motif) indicates GTPase binding domain; SM indicates SCAR-motif; WM indicates WASP-motif; VM indicates verprolin-motif; EVH1 indicates an ena/VASP homology 1 domain; EVH2 indicates an ena/VASP homology 2 domain, and FH2 indicates a Formin homology 2 domain.
Figure 5
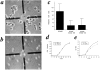
SCAR gene disruption causes reduction in cell size, but does not affect growth rate. HPS400/SCAR+ (a) and HPS400/SCAR− (b) cells were removed from suspension culture and immediately viewed by videomicroscopy. By quantitating the cell area from images such as a and b using the public domain software, NIH Image, an approximation of relative cell size is derived (c). SCAR− clones 3B and 5A are independently derived HPS400/SCAR− transformants. Error bars represent SD. (d and e) Growth curves of WT and SCAR−clones. Growth rates in terms of cell numbers are indistinguishable, but SCAR− cells show a consistent reduction of turbidity at 660 nm. Both curves are representative experiments.
Figure 6
Vegetative SCAR− cells have reduced F-actin and increased G-actin staining. F-actin stain is TRITC-phalloidin, and G-actin staining is Oregon Green 488-DNaseI. Images are three equivalent thickness confocal sections from the middle of the cells collapsed into one image. Bar, 20 μm.
Figure 7
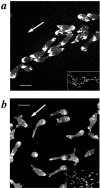
Aggregating SCAR− cells have aberrant cell morphology and F-actin staining. Aggregating cells were prepared by the submerged culture aggregation method and stained with TRITC-phalloidin. (a) HPS400/SCAR+, (b) HPS400/SCAR− cells. The white arrow represents the direction of movement; Bar, 10 μm. Inserts in a and b represent low-magnification views to more clearly compare the aggregation streams produced by the two cell lines.
Similar articles
- Control of SCAR activity in Dictyostelium discoideum.
Blagg SL, Insall RH. Blagg SL, et al. Biochem Soc Trans. 2004 Dec;32(Pt 6):1113-4. doi: 10.1042/BST0321113. Biochem Soc Trans. 2004. PMID: 15506982 - WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac.
Miki H, Suetsugu S, Takenawa T. Miki H, et al. EMBO J. 1998 Dec 1;17(23):6932-41. doi: 10.1093/emboj/17.23.6932. EMBO J. 1998. PMID: 9843499 Free PMC article. - The WASp-like protein scar regulates macropinocytosis, phagocytosis and endosomal membrane flow in Dictyostelium.
Seastone DJ, Harris E, Temesvari LA, Bear JE, Saxe CL, Cardelli J. Seastone DJ, et al. J Cell Sci. 2001 Jul;114(Pt 14):2673-83. doi: 10.1242/jcs.114.14.2673. J Cell Sci. 2001. PMID: 11683394 - Wiskott-Aldrich syndrome (role of WASP).
Nonoyama S. Nonoyama S. J Med Dent Sci. 2001 Mar;48(1):1-6. J Med Dent Sci. 2001. PMID: 12160237 Review. - The Wiskott-Aldrich syndrome protein (WASP): roles in signaling and cytoskeletal organization.
Snapper SB, Rosen FS. Snapper SB, et al. Annu Rev Immunol. 1999;17:905-29. doi: 10.1146/annurev.immunol.17.1.905. Annu Rev Immunol. 1999. PMID: 10358777 Review.
Cited by
- Hem-1 complexes are essential for Rac activation, actin polymerization, and myosin regulation during neutrophil chemotaxis.
Weiner OD, Rentel MC, Ott A, Brown GE, Jedrychowski M, Yaffe MB, Gygi SP, Cantley LC, Bourne HR, Kirschner MW. Weiner OD, et al. PLoS Biol. 2006 Feb;4(2):e38. doi: 10.1371/journal.pbio.0040038. Epub 2006 Jan 24. PLoS Biol. 2006. PMID: 16417406 Free PMC article. - Moving toward molecular mechanisms for chemotaxis in eukaryotic cells.
Devreotes P. Devreotes P. Mol Biol Cell. 2019 Nov 1;30(23):2873-2877. doi: 10.1091/mbc.E19-07-0393. Mol Biol Cell. 2019. PMID: 31671039 Free PMC article. - WASP integrates substrate topology and cell polarity to guide neutrophil migration.
Brunetti RM, Kockelkoren G, Raghavan P, Bell GRR, Britain D, Puri N, Collins SR, Leonetti MD, Stamou D, Weiner OD. Brunetti RM, et al. J Cell Biol. 2022 Feb 7;221(2):e202104046. doi: 10.1083/jcb.202104046. Epub 2021 Dec 29. J Cell Biol. 2022. PMID: 34964841 Free PMC article. - WIP-YAP/TAZ as A New Pro-Oncogenic Pathway in Glioma.
Rivas S, Antón IM, Wandosell F. Rivas S, et al. Cancers (Basel). 2018 Jun 9;10(6):191. doi: 10.3390/cancers10060191. Cancers (Basel). 2018. PMID: 29890731 Free PMC article. - Function and regulation of the Arp2/3 complex during cell migration in diverse environments.
Swaney KF, Li R. Swaney KF, et al. Curr Opin Cell Biol. 2016 Oct;42:63-72. doi: 10.1016/j.ceb.2016.04.005. Epub 2016 May 8. Curr Opin Cell Biol. 2016. PMID: 27164504 Free PMC article. Review.
References
- Adachi H, Hasebe T, Yoshinaga K, Ohta T, Sutoh K. Isolation of Dictyostelium discoideumcytokinesis mutants by restriction enzyme-mediated integration of the Blasticidin S resistence marker. Biochem Biophys Res Comm. 1994;205:1808–1814. - PubMed
- Aspenström P, Lindberg U, Hall A. Two GTPases, Cdc42 and Rac, bind directly to a protein implicated in the immunodeficiency disorder Wiskott-Aldrich syndrome. Curr Biol. 1996;6:70–75. - PubMed
- Bardwell L, Cook JG, Inouye CJ, Thorner J. Signal propagation and regulation in the mating pheromone response pathway of the yeast Saccharomyces cerevisiae. . Dev Biol. 1994;166:363–379. - PubMed
- Berks M, Kay RR. Combinatorial control of cell differentiation by cAMP and DIF-1 during development of Dictyostelium discoideum. . Development. 1990;110:977–984. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous